A specific protein substrate for a deubiquitinating enzyme: Liquid facets is the substrate of Fat facets - PubMed (original) (raw)
Comparative Study
. 2002 Feb 1;16(3):289-94.
doi: 10.1101/gad.961502.
Affiliations
- PMID: 11825870
- PMCID: PMC155328
- DOI: 10.1101/gad.961502
Comparative Study
A specific protein substrate for a deubiquitinating enzyme: Liquid facets is the substrate of Fat facets
Xin Chen et al. Genes Dev. 2002.
Abstract
Eukaryotic genomes encode large families of deubiquitinating enzymes (DUBs). Genetic data suggest that Fat facets (Faf), a Drosophila DUB essential for patterning the compound eye, might have a novel regulatory function; Faf might reverse the ubiquitination of a specific substrate, thereby preventing proteasomal degradation of that protein. Additional genetic data implicate Liquid facets (Lqf), a homolog of the vertebrate endocytic protein epsin, as a candidate for the key substrate of Faf. Here, biochemical experiments critical to testing this model were performed. The results show definitively that Lqf is the key substrate of Faf in the eye; Lqf concentration is Faf-dependent, Lqf is ubiquitinated in vivo and deubiquitinated by Faf, and Lqf and Faf interact physically.
Figures
Figure 1
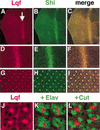
Colocalization of Shi and Lqf proteins in eye discs. (A_–_J) Apical views of a third instar larval eye disc, double-labeled with anti-Shi and anti-Lqf. (A_–_C) Lqf and Shi colocalize in cells within and posterior to the morphogenetic furrow, indicated by the arrow in A. (D_–_F) An enlargement of the area near the furrow shows that Lqf and Shi are concentrated at the apical tips of cells within the furrow. (G_–_I) An enlarged view of the area posterior to the furrow shows that Lqf and Shi are concentrated in dots, which are the apical membranes of the photoreceptors (R-cells), where they meet (see below). In addition, Lqf and Shi colocalize in a lattice, which is made up of the membranes of the surrounding cells. (J) A further enlargement of G, showing Lqf membrane localization in four adjacent facets. (K,L) An apical view of four adjacent facets in eye discs double-labeled with anti-Lqf and anti-Elav (K), which labels R-cell nuclei, or anti-Lqf and anti-Cut (L), which labels cone cell nuclei. There is no overlap in the localization of Lqf protein and either Elav or Cut; Lqf is outside the nuclei (which fill the apical cytoplasm) and in the central region of the developing facet where the photoreceptor cell membranes meet.
Figure 2
Detection of Lqf and Shi proteins in _faf_− clones in eye disc. Apical views of two different third instar larval eye discs are shown in A_–_D and E_–_H. (A,E) The _faf_− clones are labeled by the absence of β-gal protein. (B,F) The clone shapes are apparent as areas with lower levels of Lqf protein. (C,G) The clone shapes in A and E were outlined in white and layered over the panels in B and F. (D,H) The levels of Shi protein are unaffected in the _faf_− clones. We know that detection of the Lqf protein signal is unaffected by the β-gal protein signal, as clones are visible as areas of lower levels of Lqf signal in discs labeled only with anti-Lqf. A few of the clone areas in A and E are not obviously mirrored in B and F. This is because of the subtlety of the Lqf concentration difference (<twofold) often being detected; although there is only two- to threefold less Lqf in _faf_−/_faf_− eye discs than in wild-type (faf+/faf+), the clones are _faf_−/_faf_−, but the surrounding cells are often _faf_−/faf+ (the clone twin spots are faf+/faf+). Slight variability in antibody penetration and so on within the disc can affect the staining and obscure concentration differences in parts of the disc. Nevertheless, it is clear that the clone shapes are generally present in the Lqf-stained discs (B,F), but not in the Shi-stained discs (D,H).
Figure 3
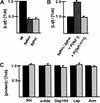
Comparison of protein levels in wild-type and _faf_− eye discs. (A) A histogram showing the level of Lqf protein, normalized to tubulin, in wild-type and _faf_− eye discs. The wild-type value was arbitrarily set to 1.0. Examples of Western blots used to generate this data are shown in Figure 4A below. (B) A histogram showing the level of Lqf protein, normalized to tubulin, in fafBX4 eye discs, and in fafBX4 eye discs with a copy of either a faf+ transgene or a _faf_− transgene. The fafBX4 value was arbitrarily set to 1.0. An example of a Western blot used to generate these data is shown in Figure 4B below. (C) A histogram showing the levels of four different endocytosis complex proteins and Armadillo (Arm), normalized to tubulin, in wild-type (wt; black bars) and fafBX4 (gray bars) eye discs. The wild-type value was arbitrarily set to 1.0. Standard errors in A_–_C were calculated from differences in repeated experiments.
Figure 4
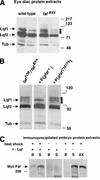
Deubiquitination and binding of Lqf by Faf. (A) Western blots of eye disc protein extracts, labeled with anti-Lqf and anti-tubulin, are shown. The lqf gene encodes two different proteins by alternate mRNA splicing, of predicted molecular weights ∼86 kD (Lqf1) and ∼70 kD (Lqf2; Cadavid et al. 2000); Lqf2 is the predominant form in eye discs. The small arrows indicate the rungs of the ladder of higher-molecular-weight forms of Lqf2. The size of the second rung of the ladder corresponds precisely to the size of Lqf1, which is the size predicted for Ub–Ub–Lqf2 (70 + 8 + 8 = 86). The two lanes shown for each genotype are different amounts of the same protein extract. These experiments were repeated 10 times, sometimes using fafFO8, and identical results were always obtained. (B) A Western blot of eye disc protein extracts, labeled with anti-Lqf and anti-tubulin. Ubiquitinated forms of Lqf2 are stabilized in _faf_− extracts, disappear when a faf+ transgene is introduced, but remain stabilized in the presence of a _faf_− transgene. Results similar to these were obtained in 3/3 repetitions. (C) Western blots of an immunoprecipitation experiment, labeled with anti-myc. The extracts are from embryos transformed with the P{hs-myc-faf} transgene that were heat-shocked and thus express myc-Faf (left and right panels), or not heat-shocked (middle panel). The extracts were immunoprecipitated with anti-Lqf (left and middle panels), or with no antibody as a control (right panel). Preimmune serum also failed to immunoprecipitate Lqf or myc-Faf (data not shown). Coomassie-stained gels of heat-shocked and non-heat-shocked embryo extracts appeared identical. The predicted molecular weight of myc-Faf is ∼300 kD. (EX) 1/15 of a 150-μL crude extract from 150 μL of embryos, (S) 1/15 of the supernatant protein from 150 μL of crude extract, which was not immunoprecipitated by anti-Lqf, (B) total protein from the 150-μL extract bound to anti-Lqf beads.
Similar articles
- Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells.
Overstreet E, Fitch E, Fischer JA. Overstreet E, et al. Development. 2004 Nov;131(21):5355-66. doi: 10.1242/dev.01434. Epub 2004 Oct 6. Development. 2004. PMID: 15469967 - In vivo Structure/Function analysis of the Drosophila fat facets deubiquitinating enzyme gene.
Chen X, Fischer JA. Chen X, et al. Genetics. 2000 Dec;156(4):1829-36. doi: 10.1093/genetics/156.4.1829. Genetics. 2000. PMID: 11102377 Free PMC article. - Either part of a Drosophila epsin protein, divided after the ENTH domain, functions in endocytosis of delta in the developing eye.
Overstreet E, Chen X, Wendland B, Fischer JA. Overstreet E, et al. Curr Biol. 2003 May 13;13(10):854-60. doi: 10.1016/s0960-9822(03)00326-9. Curr Biol. 2003. PMID: 12747835 - Endocytosis: why not wait to deubiquitinate?
Carthew RW, Xu C. Carthew RW, et al. Curr Biol. 2000 Jul 13;10(14):R532-4. doi: 10.1016/s0960-9822(00)00587-x. Curr Biol. 2000. PMID: 10898994 Review. - Protein degradation: de-ubiquitinate to decide your fate.
Kalderon D. Kalderon D. Curr Biol. 1996 Jun 1;6(6):662-5. doi: 10.1016/s0960-9822(09)00443-6. Curr Biol. 1996. PMID: 8793288 Review.
Cited by
- Caenorhabditis elegans reveals a FxNPxY-independent low-density lipoprotein receptor internalization mechanism mediated by epsin1.
Kang YL, Yochem J, Bell L, Sorensen EB, Chen L, Conner SD. Kang YL, et al. Mol Biol Cell. 2013 Feb;24(3):308-18. doi: 10.1091/mbc.E12-02-0163. Epub 2012 Dec 14. Mol Biol Cell. 2013. PMID: 23242996 Free PMC article. - The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and beta-catenin.
Murray RZ, Jolly LA, Wood SA. Murray RZ, et al. Mol Biol Cell. 2004 Apr;15(4):1591-9. doi: 10.1091/mbc.e03-08-0630. Epub 2004 Jan 23. Mol Biol Cell. 2004. PMID: 14742711 Free PMC article. - Phospho-dependent ubiquitination and degradation of PAR-1 regulates synaptic morphology and tau-mediated Aβ toxicity in Drosophila.
Lee S, Wang JW, Yu W, Lu B. Lee S, et al. Nat Commun. 2012;3:1312. doi: 10.1038/ncomms2278. Nat Commun. 2012. PMID: 23271647 Free PMC article.
References
- Baker RT, Tobias JW, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae: Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 1992;267:23364–23375. - PubMed
- Cadavid ALM. “Identification, cloning and characterization of the Drosophila liquid facets gene.” Ph.D. thesis. Austin, TX: The University of Texas at Austin; 2000.
- Cadavid ALM, Ginzel A, Fischer JA. The function of the Drosophila Fat facets deubiquitinating enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development. 2000;127:1727–1736. - PubMed
- Cagan RL, Kramer H, Hart AC, Zipursky SL. The Bride-of-sevenless and Sevenless interaction: Internalization of a transmembrane ligand. Cell. 1992;69:393–399. - PubMed
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394:793–797. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases