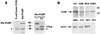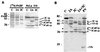Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease - PubMed (original) (raw)
Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease
N Muge Kuyumcu-Martinez et al. J Virol. 2002 Mar.
Abstract
Poliovirus (PV) causes a rapid and drastic inhibition of host cell cap-dependent protein synthesis during infection while preferentially allowing cap-independent translation of its own genomic RNA via an internal ribosome entry site element. Inhibition of cap-dependent translation is partly mediated by cleavage of an essential translation initiation factor, eIF4GI, during PV infection. In addition to cleavage of eIF4GI, cleavage of eIF4GII and poly(A)-binding protein (PABP) has been recently proposed to contribute to complete host translation shutoff; however, the relative importance of eIF4GII and PABP cleavage has not been determined. At times when cap-dependent translation is first blocked during infection, only 25 to 35% of the total cellular PABP is cleaved; therefore, we hypothesized that the pool of PABP associated with polysomes may be preferentially targeted by viral proteases. We have investigated what cleavage products of PABP are produced in vivo and the substrate determinants for cleavage of PABP by 2A protease (2A(pro)) or 3C protease (3C(pro)). Our results show that PABP in ribosome-enriched fractions is preferentially cleaved in vitro and in vivo compared to PABP in other fractions. Furthermore, we have identified four N-terminal PABP cleavage products produced during PV infection and have shown that viral 3C protease generates three of the four cleavage products. Also, 3C(pro) is more efficient in cleaving PABP in ribosome-enriched fractions than 2A(pro) in vitro. In addition, binding of PABP to poly(A) RNA stimulates 3C(pro)-mediated cleavage and inhibits 2A(pro)-mediated cleavage. These results suggest that 3C(pro) plays a major role in processing PABP during virus infection and that the interaction of PABP with translation initiation factors, ribosomes, or poly(A) RNA may promote its cleavage by viral 2A and 3C proteases.
Figures
FIG. 1.
Specificity of anti-PABP antiserum and subcellular distribution of PABP in HeLa cytoplasmic extracts. (A) (Left) Cytoplasmic extract from control bacteria (S10) or bacteria expressing recombinant His-tagged PABP was immunoblotted with rabbit polyclonal anti-PABP antiserum, and proteins were visualized by chemiluminescence. (Right) His-PABP incubated alone or with recombinant CVB3 2Apro was also immunoblotted as described above. Asterisks indicate a major cross-reactive bacterial protein. (B) HeLa cell lysates (S10) were fractionated into non-ribosome-associated (S200), crude initiation factor (RSW), and ribosome fractions and analyzed by SDS-10% PAGE followed by immunoblotting with polyclonal PABP antiserum or eIF4GI antiserum. The percentages of PABP detected in each fraction are the averages of two independent experiments. Numbers on the right show the migration of molecular weight markers (in thousands).
FIG. 2.
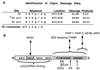
Identification of 3Cpro cleavage sites on PABP. (A) Sequences and locations of PABP cleavage sites. 3C and 3Calt sites were determined by microsequence analysis of purified cleavage products. The 3Calt" site was deduced based on gel migration of products and matching with possible 3Cpro recognition site sequences at the appropriate location in the PABP amino acid sequence. (B) Schematic diagram of the cleavage sites of viral proteases on PABP and fragments released and known binding partners for N-terminal or C-terminal domains of PABP.
FIG. 3.
Reactivity of antiserum with PABP cleavage products. (A) Radiolabeled PABP produced in vitro or HeLa cytoplasmic extract (S10) was incubated alone (C) or with 2Apro or 3Cpro. Proteins were separated on the same polyacrylamide gel, transferred to nitrocellulose, and autoradiographed (left) or immunoblotted (right) using ECL. (B) HeLa cell extract was incubated alone or with viral proteases and compared to extract from PV-infected cells that were infected at an MOI of 3 for 6 h before preparation of extract. Immunoblot detection of bands was accomplished with enhanced chemiluminescence. Asterisks indicate HeLa proteins that cross-react with PABP antiserum. Numbers on the left show the migration of molecular weight markers (in thousands).
FIG. 4.
Comparison of PABP cleavage in CVB3-infected and PV-infected HeLa cells. HeLa cells were infected with CVB3 (15 TCID50/cell) or PV (10 PFU/cell), and cytoplasmic extracts were prepared at the indicated time points. Cleavage of PABP and generation of cleavage products were detected by immunoblot and bands were visualized by 4-chloro-naphthol staining. Migration of molecular weight standards (in thousands) is indicated on the left, and major PABP cleavage products are indicated on the right.
FIG. 5.
Initiation factor- and ribosome-associated PABP is specifically targeted by enteroviral 2A and 3C proteases in vitro. HeLa S3 cells were fractionated into S200, RSW, and ribosome-enriched (RIBO) fractions. In vitro cleavage reaction mixtures (60 μl) were assembled as described in Materials and Methods by using 1 μg each of purified recombinant CVB3 2A or PV 3C protease. Cleavage reaction mixtures were incubated for 3.5 h at 37°C and analyzed by immunoblotting. The gray arrowhead indicates the putative alternate 2Apro cleavage product. Numbers on the right show the migration of molecular weight markers (in thousands).
FIG. 6.
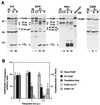
Ribosome-associated PABP is preferentially cleaved during PV infection. (A) Enhanced cleavage of ribosome-associated PABP. HeLa S3 cells were mock infected or infected with PV (MOI = 3) and harvested at the indicated time points. Harvested HeLa cells were fractionated into S200, RSW, and ribosome-enriched fractions. Aliquots of these fractions were subjected to SDS-PAGE and immunoblotted with anti-PABP serum. The percent reduction of intact PABP (percent cleavage) relative to controls is indicated below the panels. Numbers on the left show the migration of molecular weight markers (in thousands). (B) Correlation of cleavage of ribosome-associated PABP with translation shutoff. Additional HeLa infections were performed, cells were pulse-labeled with [35S]methionine, and cell lysates were analyzed by SDS-PAGE with autoradiography or immunoblotting. Host translation levels were quantitated from three sections of autoradiographs not containing viral proteins. PABP- and eIF4GI-specific immunoblotting was also performed, blots were scanned, and data were quantitated. Levels of intact PABP in mock- and PV-infected cells and levels of 2A cleavage product (CP) and C-terminal cleavage product (CPc) released by 3Cpro cleavage were determined. Levels of intact eIF4GI in cell lysates were determined by immunoblot analysis to be 100, 7.7, and 0% of mock control at 2, 3, and 4.5 h p.i., respectively. Densitometry of scanned data was performed with NIH Image 1.62. Three separate experiments are represented.
FIG. 7.
Ribosome-associated PABP is cleaved more efficiently by 3C protease than 2Apro in vitro. In vitro cleavage reaction mixtures containing increasing concentrations of 2A and 3C proteases (0.3, 1, and 3 μg/ml) were incubated with the HeLa ribosome fraction for 3.5 h at 37°C and analyzed by immunoblotting with PABP antibody. Positions of PABP cleavage products (cp) are indicated. C, control ribosome fraction incubated alone. The gray arrow represents the secondary or alternate 2Apro cleavage product observed only in vitro. Numbers on the right show the migration of molecular weight markers (in thousands).
FIG. 8.
Poly(A)-RNA-bound PABP is cleaved preferentially by 3Cpro. (A) PABP synthesized in reticulocyte lysate was separated into poly(A)-bound (B) and unbound (U) fractions as described in Materials and Methods. Unbound PABP was subjected to additional rounds of binding reactions (numbered). (B) Fractions enriched for poly(A)-bound (B) or unbound (U) PABP were incubated independently or simultaneously with both viral proteases. Cleavage reactions were analyzed by SDS-PAGE and autoradiography. (C) Products of cleavage reactions of enriched radiolabeled PABP fractions containing increasing concentrations of 2A and 3C proteases were analyzed by SDS-PAGE and autoradiography. Protease concentrations ranged from 0.3 to 3 μg/ml. The positions of the 3C, 2A, and 3Cpro alt cleavage products are indicated. Radiolabeled PABP incubated alone at 37°C in the absence of proteases served as a control (C). Numbers below panels indicate percent PABP cleaved in each reaction.
FIG. 9.
Schematic of proposed configuration of free and poly(A)-bound PABP and preferred substrates for viral proteases. PABP is depicted with four globular RRM domains and an extended flexible C-terminal tail containing a highly conserved globular domain at its tip (C terminus). RRM 2, which binds eIF4G, is hatched. Directional arrangement of poly(A) RNA-bound PABP was determined by X-ray structure (21). The CTD is shown interacting with neighboring PABP, although the actual site of binding interaction is not known. Arrows indicate enterovirus protease sites mapped on PABP, and arrow thickness shows relative cleavage efficiency. Binding of the CTD of PABP to adjacent PABP may sterically block binding of eIF4GI to all but the 3"-terminal PABP moiety. The CTD of 3"-terminal PABP is in a unique configuration and may be free to interact with the 60S ribosome (binding site hatched), eIF4B, PAIP-1, PAIP-2, and eRF3 (14, 42).
Similar articles
- Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff.
Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. Kuyumcu-Martinez NM, et al. Mol Cell Biol. 2004 Feb;24(4):1779-90. doi: 10.1128/MCB.24.4.1779-1790.2004. Mol Cell Biol. 2004. PMID: 14749392 Free PMC article. - Modulation of enteroviral proteinase cleavage of poly(A)-binding protein (PABP) by conformation and PABP-associated factors.
Rivera CI, Lloyd RE. Rivera CI, et al. Virology. 2008 May 25;375(1):59-72. doi: 10.1016/j.virol.2008.02.002. Epub 2008 Mar 5. Virology. 2008. PMID: 18321554 Free PMC article. - Cleavage of poly(A)-binding protein by poliovirus 3C proteinase inhibits viral internal ribosome entry site-mediated translation.
Bonderoff JM, Larey JL, Lloyd RE. Bonderoff JM, et al. J Virol. 2008 Oct;82(19):9389-99. doi: 10.1128/JVI.00006-08. Epub 2008 Jul 16. J Virol. 2008. PMID: 18632855 Free PMC article. - The interaction of cytoplasmic RNA viruses with the nucleus.
Weidman MK, Sharma R, Raychaudhuri S, Kundu P, Tsai W, Dasgupta A. Weidman MK, et al. Virus Res. 2003 Sep;95(1-2):75-85. doi: 10.1016/s0168-1702(03)00164-3. Virus Res. 2003. PMID: 12921997 Review. - Picornavirus 3C Proteins Intervene in Host Cell Processes through Proteolysis and Interactions with RNA.
Mondal S, Sarvari G, Boehr DD. Mondal S, et al. Viruses. 2023 Dec 12;15(12):2413. doi: 10.3390/v15122413. Viruses. 2023. PMID: 38140654 Free PMC article. Review.
Cited by
- Heat Shock Protein A6, a Novel HSP70, Is Induced During Enterovirus A71 Infection to Facilitate Internal Ribosomal Entry Site-Mediated Translation.
Su YS, Hwang LH, Chen CJ. Su YS, et al. Front Microbiol. 2021 May 7;12:664955. doi: 10.3389/fmicb.2021.664955. eCollection 2021. Front Microbiol. 2021. PMID: 34025620 Free PMC article. - Hijacking the translation apparatus by RNA viruses.
Bushell M, Sarnow P. Bushell M, et al. J Cell Biol. 2002 Aug 5;158(3):395-9. doi: 10.1083/jcb.200205044. Epub 2002 Aug 5. J Cell Biol. 2002. PMID: 12163463 Free PMC article. Review. - Translational control by viral proteinases.
Lloyd RE. Lloyd RE. Virus Res. 2006 Jul;119(1):76-88. doi: 10.1016/j.virusres.2005.10.016. Epub 2005 Nov 21. Virus Res. 2006. PMID: 16303201 Free PMC article. Review. - An update on enterovirus 71 infection and interferon type I response.
Rasti M, Khanbabaei H, Teimoori A. Rasti M, et al. Rev Med Virol. 2019 Jan;29(1):e2016. doi: 10.1002/rmv.2016. Epub 2018 Oct 30. Rev Med Virol. 2019. PMID: 30378208 Free PMC article. Review. - Nuclear translocation and regulation of intranuclear distribution of cytoplasmic poly(A)-binding protein are distinct processes mediated by two Epstein Barr virus proteins.
Park R, El-Guindy A, Heston L, Lin SF, Yu KP, Nagy M, Borah S, Delecluse HJ, Steitz J, Miller G. Park R, et al. PLoS One. 2014 Apr 4;9(4):e92593. doi: 10.1371/journal.pone.0092593. eCollection 2014. PLoS One. 2014. PMID: 24705134 Free PMC article.
References
- Asselbergs, F., W. Peters, W. van Venrooij, and H. Bloemendal. 1978. Diminished sensitivity of re-initiation of translation to inhibition by cap analogues in reticulocyte lysates. Eur. J. Biochem. 88:483-488. - PubMed
- Bag, J., and J. Wu. 1996. Translational control of poly(A)-binding protein expression. Eur. J. Biochem. 237:143-152. - PubMed
- Baker, E. J. 1993. Control of poly(A) length, p. 367-415. In J. G. Belasco and G. Brawerman (ed.), Control of messenger RNA stability. Academic Press, San Diego, Calif.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous