The S4 genome segment of baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein - PubMed (original) (raw)
The S4 genome segment of baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein
Sandra Dawe et al. J Virol. 2002 Mar.
Abstract
We demonstrate that the S4 genome segment of baboon reovirus (BRV) contains two sequential partially overlapping open reading frames (ORFs), both of which are functional in vitro and in virus-infected cells. The 15-kDa gene product (p15) of the 5"-proximal ORF induces efficient cell-cell fusion when expressed by itself in transfected cells, suggesting that p15 is the only viral protein required for induction of syncytium formation by BRV. The p15 protein is a small, hydrophobic, basic, integral membrane protein, properties shared with the p10 fusion-associated small transmembrane (FAST) proteins encoded by avian reovirus and Nelson Bay reovirus. As with p10, the BRV p15 protein is also a nonstructural protein and, therefore, is not involved in virus entry. Sequence analysis indicates that p15 shares no significant sequence similarity with the p10 FAST proteins and contains a unique repertoire and arrangement of sequence-predicted structural and functional motifs. These motifs include a functional N-terminal myristylation consensus sequence, an N-proximal proline-rich motif, two potential transmembrane domains, and an intervening polybasic region. The unique structural properties of p15 suggest that this protein is a novel member of the new family of FAST proteins.
Figures
FIG. 1.
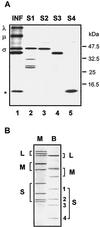
(A) Identification of the σ-class proteins encoded by the BRV S-class genome segments. The relative gel mobilities of the in vitro translation products of the BRV S1, S2, S3, and S4 genome segments, resolved by SDS-15% PAGE, are compared to the dominant viral bands present in [3H]leucine-labeled BRV-infected cell lysate (INF). The positions of the λ-, μ-, and σ-class reovirus proteins are shown at the left. The asterisk denotes the location of a ca. 15-kDa polypeptide present in the infected cell lysate that comigrates with the translation product(s) of the S4 genome segment. The relative mobilities of molecular mass markers are indicated at the right. (B) Double-stranded RNA genome segment profiles of mammalian reovirus type 3, Dearing (M), and baboon reovirus (B) resolved by SDS-10% PAGE and detected by silver staining. The positions of the BRV S1 (1,311 nt), S2 (1,253 nt), S3 (1,150 nt), and S4 genome segments are indicated on the right.
FIG. 2.
The BRV S4 genome segment contains two partially overlapping ORFs. The cDNA sequence of the BRV S4 genome segment is shown along with the predicted amino acid sequences encoded by two sequential ORFs. ORF1 (nt 25 to 447) encodes p15, a 140-amino acid protein with a predicted molecular mass of 15 kDa. ORF2 (nt 413 to 835) encodes p16, a 140- to 141-amino-acid protein with a predicted molecular mass of 16 kDa. The locations of the p15 start codon, the two adjacent potential p16 start codons, and the other two methionine codons that precede the p16 ORF, are underlined. The nucleotides comprising the 5"- and 3"-terminal noncoding regions are in lowercase. Nucleotide and amino acid numbering is indicated at the right.
FIG. 3.
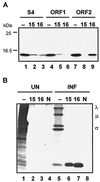
The BRV S4 genome segment is functionally bicistronic. (A) In vitro translation products of the entire S4 genome segment (lanes 1 to 3), S4 ORF1 (lanes 4 to 6), or S4 ORF2 (lanes 7 to 9) were resolved by SDS-15% PAGE and detected by fluorography. Translation reactions were fractionated without prior immunoprecipitation (−) or after immunoprecipitation with polyclonal antiserum raised against the product of ORF1, p15 (15), or the product of ORF2, p16 (16). The relative mobilities of molecular mass markers are indicated on the left. (B) [3H]leucine-labeled uninfected (UN) and BRV-infected (INF) cell lysates were fractionated by SDS-15% PAGE either without prior immune precipitation (−) or after immunoprecipitation with anti-p15 serum (15), anti-p16 serum (16), or normal rabbit serum (N). The positions of the λ-, μ-, and σ-class reovirus proteins are indicated on the right.
FIG. 4.
ORF1 of the S4 genome segment encodes the membrane fusion-inducing protein of BRV. Vero cells were transfected with pcDNA3 (a), pcDNA3-S4 (b), pcDNA3-S4 ORF1 (c), or pcDNA3-S4 ORF2 (d). Transfected monolayers were Wright-Giemsa stained 18 h posttransfection to detect the formation of multinucleated syncytia.
FIG. 5.
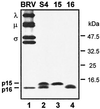
BRV p15 is a nonstructural viral protein. The relative gel mobilities of the radiolabeled in vitro translation products of BRV S4, S4 ORF1 (15), and S4 ORF2 (16) were resolved by Tricine-SDS-16.5% PAGE and compared to the viral bands present in concentrated [3H]leucine-labeled BRV particles. The positions of the λ-, μ-, and σ-class reovirus proteins and of the p15 and p16 proteins encoded by the BRV S4 genome segment are shown on the left. The migration of molecular mass markers is indicated on the right.
FIG. 6.
Sequence-predicted structural motifs and hydropathy profile of p15. The amino acid sequence of p15 is presented in the top panel, along with the locations of several predicted structural motifs, including a myristylation consensus sequence (myr), a polyproline helix (pro), a basic region, and two predicted TM domains. The bottom panel shows the hydropathy profile of p15 as determined by using the hydrophobicity scale of Kyte and Doolittle, averaged over a window of 11 residues. Regions above the horizontal line are hydrophobic, while those below the line are hydrophilic.
FIG. 7.
BRV p15 is a myristylated integral membrane protein. (A) BRV p15 was translated in vitro without or with microsomal membranes (−M or +M) in the presence of [3H]leucine. The translation products were separated into the soluble fraction (S) and the insoluble membrane fraction (I) by centrifugation. Membrane fractions were subsequently extracted with a high-pH buffer (pH) to removal peripheral membrane-associated proteins and refractionated into the soluble and insoluble fractions by centrifugation. (B) BRV-infected cells were radiolabeled with [3H]leucine, and cell lysates were fractionated into the total (T), soluble (S), and insoluble membrane (I) fractions. Samples were resolved by SDS-PAGE either without prior immunoprecipitation (−RIP) or after immunoprecipitation with normal preimmune rabbit serum (N) or anti-p15 rabbit immune serum (15). The locations of the major μ and σ proteins of BRV are indicated on the left, and the location of p15 detected by immunoprecipitation is indicated on the right. The arrow indicates a faint polypeptide present in the total and insoluble fractions that comigrates with p15. (C) Uninfected (UN) and BRV-infected (INF) [3H]myristate-labeled cell lysates were resolved by SDS-15% PAGE either without prior immunoprecipitatation (−) or after immunoprecipitatation with anti-p15 serum (15). The positions of the myristylated μ-class reovirus protein and p15 are shown on the right. The locations of molecular mass markers are indicated on the left.
FIG. 8.
Sequence-predicted structural motifs in BRV p15 and the p10 proteins of ARV and NBV. The linear arrangement, drawn to approximate scale, of sequence-predicted structural motifs of the BRV p15 protein and the p10 proteins of ARV and NBV are shown. The amino acid positions are indicated at the bottom of the figure. TM, TM domain; basic, positively charged region; m, myristylation site; PP, polyproline helix; H, hydrophobic peptide; CR, conserved motif present in ARV and NBV p10; C, conserved cysteine residues present in ARV and NBV p10 (the dicysteine motif adjacent to the TM domain in p10 is the site of palmitylation of the protein).
Similar articles
- Identification and characterization of a baboon reovirus-specific nonstructural protein encoded by the bicistronic s4 genome segment.
Dawe S, Boutilier J, Duncan R. Dawe S, et al. Virology. 2002 Dec 5;304(1):44-52. doi: 10.1006/viro.2002.1725. Virology. 2002. PMID: 12490402 - Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: implications for translation initiation.
Shmulevitz M, Yameen Z, Dawe S, Shou J, O'Hara D, Holmes I, Duncan R. Shmulevitz M, et al. J Virol. 2002 Jan;76(2):609-18. doi: 10.1128/jvi.76.2.609-618.2002. J Virol. 2002. PMID: 11752152 Free PMC article. - Atypical fusion peptide of Nelson Bay virus fusion-associated small transmembrane protein.
Cheng LT, Plemper RK, Compans RW. Cheng LT, et al. J Virol. 2005 Feb;79(3):1853-60. doi: 10.1128/JVI.79.3.1853-1860.2005. J Virol. 2005. PMID: 15650209 Free PMC article. - Reovirus FAST proteins: virus-encoded cellular fusogens.
Ciechonska M, Duncan R. Ciechonska M, et al. Trends Microbiol. 2014 Dec;22(12):715-24. doi: 10.1016/j.tim.2014.08.005. Epub 2014 Sep 19. Trends Microbiol. 2014. PMID: 25245455 Review. - Fusogenic Reoviruses and Their Fusion-Associated Small Transmembrane (FAST) Proteins.
Duncan R. Duncan R. Annu Rev Virol. 2019 Sep 29;6(1):341-363. doi: 10.1146/annurev-virology-092818-015523. Epub 2019 Jul 5. Annu Rev Virol. 2019. PMID: 31283438 Review.
Cited by
- Cell-cell membrane fusion induced by p15 fusion-associated small transmembrane (FAST) protein requires a novel fusion peptide motif containing a myristoylated polyproline type II helix.
Top D, Read JA, Dawe SJ, Syvitski RT, Duncan R. Top D, et al. J Biol Chem. 2012 Jan 27;287(5):3403-14. doi: 10.1074/jbc.M111.305268. Epub 2011 Dec 14. J Biol Chem. 2012. PMID: 22170056 Free PMC article. - Aquareovirus effects syncytiogenesis by using a novel member of the FAST protein family translated from a noncanonical translation start site.
Racine T, Hurst T, Barry C, Shou J, Kibenge F, Duncan R. Racine T, et al. J Virol. 2009 Jun;83(11):5951-5. doi: 10.1128/JVI.00171-09. Epub 2009 Mar 18. J Virol. 2009. PMID: 19297495 Free PMC article. - Unusual topological arrangement of structural motifs in the baboon reovirus fusion-associated small transmembrane protein.
Dawe S, Corcoran JA, Clancy EK, Salsman J, Duncan R. Dawe S, et al. J Virol. 2005 May;79(10):6216-26. doi: 10.1128/JVI.79.10.6216-6226.2005. J Virol. 2005. PMID: 15858006 Free PMC article. - MYRbase: analysis of genome-wide glycine myristoylation enlarges the functional spectrum of eukaryotic myristoylated proteins.
Maurer-Stroh S, Gouda M, Novatchkova M, Schleiffer A, Schneider G, Sirota FL, Wildpaner M, Hayashi N, Eisenhaber F. Maurer-Stroh S, et al. Genome Biol. 2004;5(3):R21. doi: 10.1186/gb-2004-5-3-r21. Epub 2004 Feb 13. Genome Biol. 2004. PMID: 15003124 Free PMC article. - Cell-cell fusion induced by reovirus FAST proteins enhances replication and pathogenicity of non-enveloped dsRNA viruses.
Kanai Y, Kawagishi T, Sakai Y, Nouda R, Shimojima M, Saijo M, Matsuura Y, Kobayashi T. Kanai Y, et al. PLoS Pathog. 2019 Apr 25;15(4):e1007675. doi: 10.1371/journal.ppat.1007675. eCollection 2019 Apr. PLoS Pathog. 2019. PMID: 31022290 Free PMC article.
References
- Bonner, W. M. 1984. Fluorography for the detection of radioactivity in gels. Methods Enzymol. 104:460-465. - PubMed
- Duncan, R. 1999. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology 260:316-328. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials