Phylogeographic differentiation of mitochondrial DNA in Han Chinese - PubMed (original) (raw)
Phylogeographic differentiation of mitochondrial DNA in Han Chinese
Yong-Gang Yao et al. Am J Hum Genet. 2002 Mar.
Abstract
To characterize the mitochondrial DNA (mtDNA) variation in Han Chinese from several provinces of China, we have sequenced the two hypervariable segments of the control region and the segment spanning nucleotide positions 10171-10659 of the coding region, and we have identified a number of specific coding-region mutations by direct sequencing or restriction-fragment-length-polymorphism tests. This allows us to define new haplogroups (clades of the mtDNA phylogeny) and to dissect the Han mtDNA pool on a phylogenetic basis, which is a prerequisite for any fine-grained phylogeographic analysis, the interpretation of ancient mtDNA, or future complete mtDNA sequencing efforts. Some of the haplogroups under study differ considerably in frequencies across different provinces. The southernmost provinces show more pronounced contrasts in their regional Han mtDNA pools than the central and northern provinces. These and other features of the geographical distribution of the mtDNA haplogroups observed in the Han Chinese make an initial Paleolithic colonization from south to north plausible but would suggest subsequent migration events in China that mainly proceeded from north to south and east to west. Lumping together all regional Han mtDNA pools into one fictive general mtDNA pool or choosing one or two regional Han populations to represent all Han Chinese is inappropriate for prehistoric considerations as well as for forensic purposes or medical disease studies.
Figures
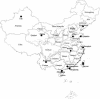
Figure 1
Geographic locations of the Han samples under study
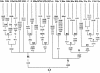
Figure 2
Classification tree of the mtDNA haplogroups observed in Han Chinese. The diagnostic mutations considered here (relative to the revised CRS; Andrews et al. 1999) are indicated on the branches. Nucleotide changes are specified for transversions by suffixes, and “d” indicates deletion; mutations recurrent in this tree are underlined. The revised CRS differs from the root of haplogroup R by mutations at 73, 2706, 7028, 11719, and 14766 that are characteristic of HV or H and by seven private mutations at 263, 315+C, 750, 1438, 4769, 8860, and 15326 (Andrews et al. 1999).
Figure 3
PC map of the mtDNA data (with respect to the basal haplogroup profiles) of 13 regional Han samples.
Similar articles
- Analysis of mitochondrial DNA polymorphisms in Guangdong Han Chinese.
Chen F, Wang SY, Zhang RZ, Hu YH, Gao GF, Liu YH, Kong QP. Chen F, et al. Forensic Sci Int Genet. 2008 Mar;2(2):150-3. doi: 10.1016/j.fsigen.2007.10.122. Forensic Sci Int Genet. 2008. PMID: 19083810 - Population data of mitochondrial DNA HVS-I and HVS-II sequences for 208 Henan Han Chinese.
Xu K, Hu S. Xu K, et al. Leg Med (Tokyo). 2015 Jul;17(4):287-94. doi: 10.1016/j.legalmed.2015.02.003. Epub 2015 Feb 24. Leg Med (Tokyo). 2015. PMID: 25759193 - Phylogeographic analysis of mtDNA variation in four ethnic populations from Yunnan Province: new data and a reappraisal.
Yao YG, Zhang YP. Yao YG, et al. J Hum Genet. 2002;47(6):311-8. doi: 10.1007/s100380200042. J Hum Genet. 2002. PMID: 12111379 - Phylogeographic analysis of mitochondrial DNA in northern Asian populations.
Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Dambueva I, Perkova M, Dorzhu C, Luzina F, Lee HK, Vanecek T, Villems R, Zakharov I. Derenko M, et al. Am J Hum Genet. 2007 Nov;81(5):1025-41. doi: 10.1086/522933. Epub 2007 Oct 1. Am J Hum Genet. 2007. PMID: 17924343 Free PMC article. - Ancient DNA reveals genetic connections between early Di-Qiang and Han Chinese.
Li J, Zeng W, Zhang Y, Ko AM, Li C, Zhu H, Fu Q, Zhou H. Li J, et al. BMC Evol Biol. 2017 Dec 4;17(1):239. doi: 10.1186/s12862-017-1082-0. BMC Evol Biol. 2017. PMID: 29202706 Free PMC article.
Cited by
- Evidence of ancient DNA reveals the first European lineage in Iron Age Central China.
Xie CZ, Li CX, Cui YQ, Zhang QC, Fu YQ, Zhu H, Zhou H. Xie CZ, et al. Proc Biol Sci. 2007 Jul 7;274(1618):1597-601. doi: 10.1098/rspb.2007.0219. Proc Biol Sci. 2007. PMID: 17456455 Free PMC article. - HLA variation reveals genetic continuity rather than population group structure in East Asia.
Di D, Sanchez-Mazas A. Di D, et al. Immunogenetics. 2014 Mar;66(3):153-60. doi: 10.1007/s00251-014-0757-6. Epub 2014 Jan 22. Immunogenetics. 2014. PMID: 24449274 - MtDNA control region sequence polymorphisms and phylogenetic analysis of Malay population living in or around Kuala Lumpur in Malaysia.
Maruyama S, Nohira-Koike C, Minaguchi K, Nambiar P. Maruyama S, et al. Int J Legal Med. 2010 Mar;124(2):165-70. doi: 10.1007/s00414-009-0355-6. Epub 2009 Jun 16. Int J Legal Med. 2010. PMID: 19533161 - APOE gene polymorphism in long-lived individuals from a central China population.
Liu G, Liu X, Yu P, Wang Q, Wang H, Li C, Ye G, Wu X, Tan C. Liu G, et al. Sci Rep. 2017 Jun 12;7(1):3292. doi: 10.1038/s41598-017-03227-5. Sci Rep. 2017. PMID: 28607446 Free PMC article. - Discovery of a novel genetic susceptibility locus on X chromosome for systemic lupus erythematosus.
Zhu Z, Liang Z, Liany H, Yang C, Wen L, Lin Z, Sheng Y, Lin Y, Ye L, Cheng Y, Chang Y, Liu L, Yang L, Shi Y, Shen C, Zhou F, Zheng X, Zhu J, Liang B, Ding Y, Zhou Y, Yin X, Tang H, Zuo X, Sun L, Bei JX, Liu J, Yang S, Yang W, Cui Y, Zhang X. Zhu Z, et al. Arthritis Res Ther. 2015 Dec 3;17:349. doi: 10.1186/s13075-015-0857-1. Arthritis Res Ther. 2015. PMID: 26635088 Free PMC article.
References
Electronic-Database Information
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for mtDNA control region data; accession numbers AY052834–AY053358)
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 - PubMed
- Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 - PubMed
- Bandelt H-J, Macaulay V, Richards M (2000) Median networks: speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA. Mol Phylogenet Evol 16:8–28 - PubMed
- Bandelt H-J, Lahermo P, Richards M, Macaulay V (2001) Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med 115:64–69 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources