Functional and physical interactions between components of the Prp19p-associated complex - PubMed (original) (raw)
Functional and physical interactions between components of the Prp19p-associated complex
Chun-Hong Chen et al. Nucleic Acids Res. 2002.
Abstract
The Prp19p-associated complex is essential for the yeast pre-mRNA splicing reaction. The complex consists of at least eight protein components, but is not tightly associated with spliceosomal snRNAs. By a combination of genetic and biochemical methods we previously identified four components of this complex, Ntc25p, Ntc85p, Ntc30p and Ntc20p, all of them being novel splicing factors. We have now identified three other components of the complex, Ntc90p, Ntc77p and Ntc31p. These three proteins were also associated with the spliceosome during the splicing reaction in the same manner as Prp19p, concurrently with or immediately after dissociation of U4 snRNA. Two-hybrid analysis revealed that none of these proteins interacted with Prp19p or Ntc25p, but all interacted with Ntc85p. An interaction network between the identified components of the Prp19p-associated complex is demonstrated. Biochemical analysis revealed that Ntc90p, Ntc31p, Ntc30p and Ntc20p form a subcomplex, which, through interacting with Ntc85p and Ntc77p, can associate with Prp19p and Ntc25p to form the Prp19p-associated complex. Genetic analysis suggests that Ntc31p, Ntc30p and Ntc20p may play roles in modulating the function of Ntc90p.
Figures
Figure 1
The Prp19p-associated complex was isolated by affinity chromatography on an anti-HA antibody column using the PRP19-HA extract and the components were fractionated by SDS–PAGE on a 10% gel. The extract without a tag was used as the background control (–). The high molecular weight region was enlarged as shown on the left.
Figure 2
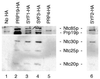
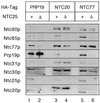
Identification of Syf1p, Syf2p and Syf3p as components of the Prp19p-associated complex. Syf1p, Syf2p and Syf3p were individually tagged with the HA epitope. Immunoprecipitation with the anti-HA antibody of extracts prepared from these strains (lanes 3, 4 and 6) revealed co-precipitation of the identified Prp19p-associated components.
Figure 3
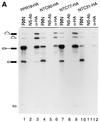
(A) Immunoprecipitation of the spliceosome formed in PRP19-HA (lanes 1–3), NTC90-HA (lanes 4–6), NTC77-HA (lanes 7–9) and NTC31-HA (lanes 10–12) extracts with the anti-HA antibody (lanes 3, 6, 9 and 12). (B) Immunoprecipitation of the spliceosome formed at various ATP concentrations in extracts prepared from a strain in which Prp19p was tagged with the c-Myc epitope and Prp4p tagged with the HA epitope. The splicing reaction was carried out at 0.5 mM (lanes 1–7), 0.1 mM (lanes 8–14) and 0.05 mM (lanes 15–21) ATP and the reaction mixtures (15 µl each) were subjected to immunoprecipitation with protein A–Sepharose alone (lanes 2, 9 and 16), anti-Myc (lanes 3, 10 and 17), anti-HA (lanes 4, 11 and 18), anti-Ntc90p (lanes 5, 12 and 19), anti-Ntc77p (lanes 6, 13 and 20) and anti-Ntc31p antibodies (lanes 7, 14 and 21). RXN, 2 µl of the reaction mixture; NS-Ab, non-specific antibody; PAS, protein A–Sepharose.
Figure 3
(A) Immunoprecipitation of the spliceosome formed in PRP19-HA (lanes 1–3), NTC90-HA (lanes 4–6), NTC77-HA (lanes 7–9) and NTC31-HA (lanes 10–12) extracts with the anti-HA antibody (lanes 3, 6, 9 and 12). (B) Immunoprecipitation of the spliceosome formed at various ATP concentrations in extracts prepared from a strain in which Prp19p was tagged with the c-Myc epitope and Prp4p tagged with the HA epitope. The splicing reaction was carried out at 0.5 mM (lanes 1–7), 0.1 mM (lanes 8–14) and 0.05 mM (lanes 15–21) ATP and the reaction mixtures (15 µl each) were subjected to immunoprecipitation with protein A–Sepharose alone (lanes 2, 9 and 16), anti-Myc (lanes 3, 10 and 17), anti-HA (lanes 4, 11 and 18), anti-Ntc90p (lanes 5, 12 and 19), anti-Ntc77p (lanes 6, 13 and 20) and anti-Ntc31p antibodies (lanes 7, 14 and 21). RXN, 2 µl of the reaction mixture; NS-Ab, non-specific antibody; PAS, protein A–Sepharose.
Figure 4
Two-hybrid assays showing interactions between the Prp19p-associated components. (A) Ntc90p was fused to the GAL4 AD and the counterparts fused to the LexA BD. (B) Ntc31p was fused to the GAL4 AD and the counterparts fused to the LexA BD. (C) Left, Ntc77p was fused to the LexA BD and the counterparts fused to the GAL4 AD. Right, Ntc77p was fused to the GAL4 AD and the counterparts fused to the GAL4 BD.
Figure 5
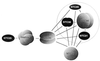
A scheme showing interactions between the components of the Prp19p-associated complex. Prp19p, Ntc90p, Ntc85p and Ntc30p are shown as multi-subunit homo-oligomers due to observed self-interactions in two-hybrid assays. Prp19p was demonstrated to be in tetrameric form. Self-interaction was not detected for Ntc77p, Ntc31p, Ntc25p or Ntc20p. Proteins encoded by essential genes are shown in gray and non-essential genes in black. The circle indicates stable association of Ntc90p, Ntc31p, Ntc30p and Ntc20p in a subcomplex.
Figure 6
Growth analysis of NTC31, NTC30 and NTC20 single, double and triple deletion mutants. (A) Growth phenotypes of single and double deletion mutants of NTC31, NTC30 and NTC20 were examined by spot assay on YPD plates. (B) Cells of the wild-type, double deletion and triple deletion strains carrying plasmid pRS416.NTC30 were streaked on YPD or 5-FOA plates. WT, wild-type.
Figure 7
The growth defect of a ΔNTC30/ΔNTC20 mutant could be partially rescued by overexpression of NTC90. (A) ΔNTC30/ΔNTC20 cells carrying plasmids containing individual genes encoding components of the Prp19p-associated complex in plasmid vector pG1 were spotted on YPD plates and grown at 30, 35 and 37°C. (B) Wild-type, ΔNTC30/ΔNTC20 or ΔNTC25 cells carrying the NTC90 gene under control of the GAL promoter on plasmid vector pRS414 were spotted on tryptophan drop-out–SMM plates supplemented with galactose or glucose and grown at 30, 35 and 37°C. WT, wild-type.
Figure 8
Immunoprecipitation of ΔNTC25 extracts revealed stable association of Ntc90p, Ntc31p, Ntc30p and Ntc20p in a subcomplex. Prp19p, Ntc20p or Ntc77p was tagged with the HA epitope in a strain in which the NTC25 gene was deleted. Extracts prepared from such strains were precipitated with anti-HA antibody followed by western blotting.
Similar articles
- Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p.
Chen CH, Tsai WY, Chen HR, Wang CH, Cheng SC. Chen CH, et al. J Biol Chem. 2001 Jan 5;276(1):488-94. doi: 10.1074/jbc.M006958200. J Biol Chem. 2001. PMID: 11018040 - Characterization of interactions among the Cef1p-Prp19p-associated splicing complex.
Ohi MD, Gould KL. Ohi MD, et al. RNA. 2002 Jun;8(6):798-815. doi: 10.1017/s1355838202025050. RNA. 2002. PMID: 12088152 Free PMC article. - Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing.
Tsai WY, Chow YT, Chen HR, Huang KT, Hong RI, Jan SP, Kuo NY, Tsao TY, Chen CH, Cheng SC. Tsai WY, et al. J Biol Chem. 1999 Apr 2;274(14):9455-62. doi: 10.1074/jbc.274.14.9455. J Biol Chem. 1999. PMID: 10092627 - Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing.
Chen HR, Tsao TY, Chen CH, Tsai WY, Her LS, Hsu MM, Cheng SC. Chen HR, et al. Proc Natl Acad Sci U S A. 1999 May 11;96(10):5406-11. doi: 10.1073/pnas.96.10.5406. Proc Natl Acad Sci U S A. 1999. PMID: 10318896 Free PMC article.
Cited by
- Ntc90 is required for recruiting first step factor Yju2 but not for spliceosome activation.
Chang KJ, Chen HC, Cheng SC. Chang KJ, et al. RNA. 2009 Sep;15(9):1729-39. doi: 10.1261/rna.1625309. Epub 2009 Jul 17. RNA. 2009. PMID: 19617314 Free PMC article. - Malleable ribonucleoprotein machine: protein intrinsic disorder in the Saccharomyces cerevisiae spliceosome.
Coelho Ribeiro Mde L, Espinosa J, Islam S, Martinez O, Thanki JJ, Mazariegos S, Nguyen T, Larina M, Xue B, Uversky VN. Coelho Ribeiro Mde L, et al. PeerJ. 2013 Feb 12;1:e2. doi: 10.7717/peerj.2. Print 2013. PeerJ. 2013. PMID: 23638354 Free PMC article. - Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions.
Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R. Deckert J, et al. Mol Cell Biol. 2006 Jul;26(14):5528-43. doi: 10.1128/MCB.00582-06. Mol Cell Biol. 2006. PMID: 16809785 Free PMC article. - The Saccharomyces cerevisiae gene CDC40/PRP17 controls cell cycle progression through splicing of the ANC1 gene.
Dahan O, Kupiec M. Dahan O, et al. Nucleic Acids Res. 2004 May 7;32(8):2529-40. doi: 10.1093/nar/gkh574. Print 2004. Nucleic Acids Res. 2004. PMID: 15133121 Free PMC article. - Thermoconditional modulation of the pleiotropic sensitivity phenotype by the Saccharomyces cerevisiae PRP19 mutant allele pso4-1.
Revers LF, Cardone JM, Bonatto D, Saffi J, Grey M, Feldmann H, Brendel M, Henriques JA. Revers LF, et al. Nucleic Acids Res. 2002 Nov 15;30(22):4993-5003. doi: 10.1093/nar/gkf632. Nucleic Acids Res. 2002. PMID: 12434004 Free PMC article.
References
- Beggs J.D. (1993) Yeast protein splicing factors involved in nuclear pre-mRNA splicing. Mol. Biol. Rep., 18, 99–103. - PubMed
- Guthrie C. (1991) Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science, 253, 157–163. - PubMed
- Moore M.J., Query,C.C. and Sharp,P.A. (1993) Splicing of precursors to mRNAs by the spliceosome. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 303–357.
- Rymond B.C. and Rosbash,M. (1992) Yeast pre-mRNA splicing. In Jones,E.W., Pringle,J.R. and Broach,R.J. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 2, pp. 143–192.
- Sharp P.A. (1994) Split genes and RNA splicing. Cell, 77, 805–815. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases