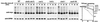Role of the ubiquitin-selective CDC48(UFD1/NPL4 )chaperone (segregase) in ERAD of OLE1 and other substrates - PubMed (original) (raw)
Role of the ubiquitin-selective CDC48(UFD1/NPL4 )chaperone (segregase) in ERAD of OLE1 and other substrates
Sigurd Braun et al. EMBO J. 2002.
Abstract
The OLE pathway of yeast regulates the abundance of the ER-bound enzyme Delta-9 fatty acid desaturase OLE1, thereby controlling unsaturated fatty acid pools and membrane fluidity. Previously, we showed that this pathway is exquisitely regulated by the ubiquitin/proteasome system. Activation of the pathway involves proteasomal processing of a membrane-bound transcription factor and the subsequent mobilization of the cleaved, ubiquitylated transcription factor from its partner molecule by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone-like enzyme. Here we report that the OLE1 protein itself is naturally short-lived and is degraded by ubiquitin/proteasome-dependent ER-associated degradation (ERAD). We found that CDC48(UFD1/NPL4) plays a second role in the OLE pathway by mediating ERAD of OLE1. Intriguingly, other ERAD substrates also require CDC48(UFD1/NPL4) for degradation, indicating that this enzyme is a novel, constitutive component of the ERAD machinery. We propose that CDC48(UFD1/NPL4) functions as a segregase that liberates ubiquitylated proteins from non-modified partners.
Figures
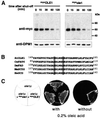
Fig. 1. OLE1 is short-lived in vivo. (A) Expression shut-off experiments with WT cells expressing mycOLE1 and the mutant variant mycole1 under the control of the GAL1-10 promoter. Cells were grown in YPGal to an OD600 of 0.5 at 23°C and shifted for another 2 h to 37°C. The experiment was started by adding glucose and cycloheximide to the medium. At each time point indicated, the cellular level of both epitope-tagged OLE1 variants was analyzed by anti-myc immunoblots (upper panel). As a control, the blots were reprobed with an antibody against the stable ER membrane protein dolichol phosphate mannose synthase, DPM1 (lower panel). (B) Sequence comparison of Δ-9 fatty acid desaturases from different organisms (Sc, Saccharomyces cerevisiae; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Mm, Mus musculus; Hs, Homo sapiens). The two conserved His residues, which were replaced by Ala residues in the mutant ole1 variant, are shaded in gray. (C) Growth of the ole1Δ deletion strain expressing either none, mycOLE1 or mutant mycole1 in the presence and absence of oleic acid, respectively. The lethal phenotype of ole1Δ in the absence of unsaturated fatty acids in the growth medium can only be suppressed by expressing the functional desaturase.
Fig. 2. OLE1 turnover is mediated by ERAD. Expression shut-off experiments with different ERAD mutants expressing the mutant mycole1 variant under the control of the GAL1-10 promoter. Experimental procedures were identical to those described in Figure 1A. Shown are the protein levels for mycole1 (upper panels) and as a control for DPM1 (lower panels). The right panel shows the quantification of the mycole1 decay. The estimated half-lives of mycole1 in WT, ubc6Δ ubc7Δ, cue1Δ, hrd1Δ, cim5-1 and sec61-R2 strains were 25, 80, 80, 35, 55 and 70 min, respectively.
Fig. 3. Involvement of the CDC48UFD1/NPL4 segregase in OLE1 degradation. (A) Expression shut-off experiments with the ts mutants ufd1-2, npl4-1, cdc48-6 and the deletion strain shp1Δ expressing the mutant mycole1 variant under the control of the GAL1-10 promoter in the absence of oleic acid (at 37°C). Shown are the protein levels for mycole1 (upper panels) and as a control for DPM1 (lower panels). The right panel shows the quantification of the mycole1 decay. The estimated half-lives of mycole1 in WT, ufd1-1, npl4-1, cdc48-6 and shp1Δ strains were 25, 275, 110, 60 and 27 min, respectively. (B) The same experiment as in (A) was performed in the presence of 0.2% oleic acid added 1 h prior to the 37°C shift to the medium. (C) Yeast two-hybrid interaction of CDC48 and SHP1. Open reading frames of SHP1 and CDC48 were cloned into pGAD424 and pGBD, respectively. Transformants were streaked onto SC-Leu-Trp-His plates to test for two-hybrid interaction. Empty vectors are indicated by ‘–’. (D) Co-immunoprecipitation of CDC48 with SHP1. Cytosolic fractions from yeast cells expressing epitope-tagged VSVCDC48 (indicated by ‘+’) and mycSHP1 were subjected to anti-myc and anti-IgG immunoprecipitation, respectively. The panel designated as ‘input’ shows 10% of the amount of protein used for immunoprecipitation.
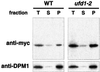
Fig. 4. Accumulation of stabilized OLE1 at the membrane. WT and ufd1-2 cells expressing mutant mycole1 under the control of the GAL1-10 promoter were grown in YPGal to a final OD600 of 1.0. An expression shut-off assay was performed by adding glucose and cycloheximide to the medium. After 30 min, cells corresponding to 50 OD600 (for both yeast strains) were subjected to cell fractionation as described in Materials and methods. Equal amounts of total extract (T), soluble (S) and pellet (P) fraction were analyzed by immunoblotting with anti-myc (upper panel) and the ER membrane protein anti-DPM1 (lower panel) antibody, respectively.
Fig. 5. General role of the CDC48UFD1/NPL4 segregase in ERAD. (A) Expression shut-off (cycloheximide chase) with WT and ufd1-2, npl4-1, cdc48-6 and the deletion strain shp1Δ expressing mycHMG2. Cells were grown in YPD to an OD600 of 0.5 at 23°C and then shifted for another 2 h to 37°C. After adding cycloheximide to the medium, samples were taken at the time points indicated and protein extracts were prepared. The protein level of epitope-tagged mycHMG2 is shown by performing western blots with an anti-myc antibody (upper panel). As a control, blots were reprobed with an antibody against the stable ER membrane protein DPM1 (lower panel). The right panel shows the quantification of the mycHMG2 decay. The estimated half-lives of mycHMG2 in WT, ufd1-1, npl4-1, cdc48-6 and shp1Δ strains were 20, 175, 110, 70 and 30 minutes, respectively. (B) Steady state level of the unstable DEG1SEC62FLAG variant in WT, ufd1-2, npl4-1, cdc48-6 cells and the deletion strain shp1Δ. Cells were grown in YPGal to an OD600 of 0.5 at 23°C and after another 2 h of growth at 37°C, aliquots were taken and protein samples prepared. The steady state level of the epitope-tagged SEC62 variant was analyzed by using an anti-FLAG antibody (upper panel). As a control, blots were reprobed with an antibody against DPM1 (lower panel).
Similar articles
- Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone.
Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Rape M, et al. Cell. 2001 Nov 30;107(5):667-77. doi: 10.1016/s0092-8674(01)00595-5. Cell. 2001. PMID: 11733065 - Cdc48-Ufd1-Npl4: stuck in the middle with Ub.
Bays NW, Hampton RY. Bays NW, et al. Curr Biol. 2002 May 14;12(10):R366-71. doi: 10.1016/s0960-9822(02)00862-x. Curr Biol. 2002. PMID: 12015140 Review. - Ssz1 restores endoplasmic reticulum-associated protein degradation in cells expressing defective cdc48-ufd1-npl4 complex by upregulating cdc48.
Bosis E, Salomon D, Ohayon O, Sivan G, Bar-Nun S, Rabinovich E. Bosis E, et al. Genetics. 2010 Mar;184(3):695-706. doi: 10.1534/genetics.109.111419. Epub 2009 Dec 28. Genetics. 2010. PMID: 20038635 Free PMC article. - Cdc48 and cofactors Npl4-Ufd1 are important for G1 progression during heat stress by maintaining cell wall integrity in Saccharomyces cerevisiae.
Hsieh MT, Chen RH. Hsieh MT, et al. PLoS One. 2011 Apr 19;6(4):e18988. doi: 10.1371/journal.pone.0018988. PLoS One. 2011. PMID: 21526151 Free PMC article. - Cdc48/p97 segregase: Spotlight on DNA-protein crosslinks.
Noireterre A, Stutz F. Noireterre A, et al. DNA Repair (Amst). 2024 Jul;139:103691. doi: 10.1016/j.dnarep.2024.103691. Epub 2024 May 9. DNA Repair (Amst). 2024. PMID: 38744091 Review.
Cited by
- ATP binding to p97/VCP D1 domain regulates selective recruitment of adaptors to its proximal N-domain.
Chia WS, Chia DX, Rao F, Bar Nun S, Geifman Shochat S. Chia WS, et al. PLoS One. 2012;7(12):e50490. doi: 10.1371/journal.pone.0050490. Epub 2012 Dec 3. PLoS One. 2012. PMID: 23226521 Free PMC article. - Monoubiquitination of EEA1 regulates endosome fusion and trafficking.
Ramanathan HN, Zhang G, Ye Y. Ramanathan HN, et al. Cell Biosci. 2013 May 23;3(1):24. doi: 10.1186/2045-3701-3-24. Cell Biosci. 2013. PMID: 23701900 Free PMC article. - VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments.
Wang Y, Satoh A, Warren G, Meyer HH. Wang Y, et al. J Cell Biol. 2004 Mar 29;164(7):973-8. doi: 10.1083/jcb.200401010. Epub 2004 Mar 22. J Cell Biol. 2004. PMID: 15037600 Free PMC article. - Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy.
Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, Dargemont C. Ossareh-Nazari B, et al. EMBO Rep. 2010 Jul;11(7):548-54. doi: 10.1038/embor.2010.74. Epub 2010 May 28. EMBO Rep. 2010. PMID: 20508643 Free PMC article. - Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity.
Duennwald ML, Lindquist S. Duennwald ML, et al. Genes Dev. 2008 Dec 1;22(23):3308-19. doi: 10.1101/gad.1673408. Epub 2008 Nov 17. Genes Dev. 2008. PMID: 19015277 Free PMC article.
References
- Bays N.W., Gardner,R.G., Seelig,L.P., Joazeiro,C.A. and Hampton,R.Y. (2001) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature Cell Biol., 3, 24–29. - PubMed
- Biederer T., Volkwein,C. and Sommer,T. (1997) Role of Cue1p in ubiquitination and degradation at the ER surface. Science, 278, 1806–1809. - PubMed
- Chen P., Johnson,P., Sommer,T., Jentsch,S. and Hochstrasser,M. (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT α2 repressor. Cell, 74, 357–369. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous