Generation of ligand-receptor alliances by "SEA" module-mediated cleavage of membrane-associated mucin proteins - PubMed (original) (raw)
. 2002 Mar;11(3):698-706.
doi: 10.1110/ps.16502.
Michael A McGuckin, Stefanie J Williams, Amos Baruch, Merav Yoeli, Ravit Ziv, Liron Okun, Joseph Zaretsky, Nechama Smorodinsky, Iafa Keydar, Pavlos Neophytou, Martin Stacey, His-Hsien Lin, Siamon Gordon
Affiliations
- PMID: 11847293
- PMCID: PMC2373471
- DOI: 10.1110/ps.16502
Generation of ligand-receptor alliances by "SEA" module-mediated cleavage of membrane-associated mucin proteins
Daniel H Wreschner et al. Protein Sci. 2002 Mar.
Abstract
A mechanism is described whereby one and the same gene can encode both a receptor protein as well as its specific ligand. Generation of this receptor-ligand partnership is effected by proteolytic cleavage within a specific module located in a membrane resident protein. It is postulated here that the "SEA" module, found in a number of heavily O-linked glycosylated membrane-associated proteins, serves as a site for proteolytic cleavage. The subunits generated by proteolytic cleavage of the SEA module reassociate, and can subsequently elicit a signaling cascade. We hypothesize that all membrane resident proteins containing such a "SEA" module will undergo cleavage, thereby generating a receptor-ligand alliance. This requires that the protein subunits resulting from the proteolytic cleavage reassociate with each other in a highly specific fashion. The same SEA module that serves as the site for proteolytic cleavage, probably also contains the binding sites for reassociation of the resultant two subunits. More than one type of module can function as a site for proteolytic cleavage; this can occur not only in one-pass membrane proteins but also in 7-transmembrane proteins and other membrane-associated proteins. The proposal presented here is likely to have significant practical consequences. It could well lead to the rational design and identification of molecules that, by binding to one of the cleaved partners, will act either as agonists or antagonists, alter signal transduction and, hence, cellular behavior.
Figures
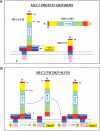
Fig. 1.
MUC1 isoforms and their interactions. (A) MUC1 isoforms. N and C refer to the amino and carboxyl termini of the proteins. Regions common to the various MUC1 isoforms have the same colours and shadings. SP and VNTR refer to the signal peptide and variable number of tandem repeats, respectively. The splice donor and acceptor sites used to generate the MUC1/Y isoform are indicated by S.D. and S.A., respectively. The membrane proximal binding site is indicated by "MEM. Site," whereas the binding site of the large extracellular domain is designated "EX site." Both are contained within the SEA module. (B) Signaling initiated by the release and reassociation of the large extracellular domain.
Fig. 2.
Across species comparison of the MUC1/TM protein. The sequences were taken from Spicer et al. (1995). The conserved YQEL and GSVVV sites, also noted and discussed in Spicer et al. (1995), are highlighted and the site of cleavage is indicated.
Fig. 3.
Conserved SEA module sequences in other proteins upstream of the GSVVV site. The MUC1 (P15941, accession numbers in brackets), MUC3 (AF143373), MUC12 (AF147790), Interphotoreceptor proteoglycan 200 (AF173155), C. elegans MUC (CAB03861) are aligned for maximal homology. The conserved LED, YQEL, and GSVVV sites are highlighted and the interdigitating hydrophilic (yellow on red background) and hydrophobic (yellow on blue background) amino acids just upstream of the cleavage site (vertical downward facing arrow) are also indicated. The small print (green) amino acids in the MUC1, MUC3, and perlecan sequences indicate the splice sites. These are also shown by the red stars. Numbers next to the stars indicate the intron phase.
Fig. 4.
C. elegans SEA module-containing transmembrane protein. (A) Sequence of C. elegans SEA module-containing transmembrane protein. Yellow letters on a blue background indicate the leader signal peptide and transmembrane domains. The transfer stop signal immediately downstream to the transmembrane domain is written in bold, italic, underlined letters, and the cytoplasmic tyrosine residues are highlighted in red. White letters on a gray background indicate the SEA module juxtaposed upstream of the transmembrane. The conserved motifs within the SEA module are indicated by red letters on a yellow background. Yellow letters on a green background show the strings of threonine and serine residues, and the potential N-glycosylation sites are highlighted yellow on a red background. (B) Comparison of C. elegans SEA module-containing transmembrane protein with human MUC1. The two sequences are aligned for maximum similarity. Conservatively substituted amino acids are indicated by a plus sign. Note that all hydrophilic amino acids (K, R, D, and E) are considered conservative substitutions for each other.
Fig. 4.
C. elegans SEA module-containing transmembrane protein. (A) Sequence of C. elegans SEA module-containing transmembrane protein. Yellow letters on a blue background indicate the leader signal peptide and transmembrane domains. The transfer stop signal immediately downstream to the transmembrane domain is written in bold, italic, underlined letters, and the cytoplasmic tyrosine residues are highlighted in red. White letters on a gray background indicate the SEA module juxtaposed upstream of the transmembrane. The conserved motifs within the SEA module are indicated by red letters on a yellow background. Yellow letters on a green background show the strings of threonine and serine residues, and the potential N-glycosylation sites are highlighted yellow on a red background. (B) Comparison of C. elegans SEA module-containing transmembrane protein with human MUC1. The two sequences are aligned for maximum similarity. Conservatively substituted amino acids are indicated by a plus sign. Note that all hydrophilic amino acids (K, R, D, and E) are considered conservative substitutions for each other.
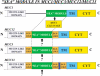
Fig. 5.
Proposed cleavage in the MUC12, MUC3, and MUC13 SEA modules. The various proteins are shown schematically. The amino-terminal barred region indicates the tandem repeat mucin domains of these proteins. TM and CYT designate the transmembrane and cytoplasmic domains. The MUC12, MUC3, and MUC13 all have EGF-like domains that bracket the SEA module.
Similar articles
- Identification of MUC1 proteolytic cleavage sites in vivo.
Parry S, Silverman HS, McDermott K, Willis A, Hollingsworth MA, Harris A. Parry S, et al. Biochem Biophys Res Commun. 2001 May 11;283(3):715-20. doi: 10.1006/bbrc.2001.4775. Biochem Biophys Res Commun. 2001. PMID: 11341784 - The MUC1 SEA module is a self-cleaving domain.
Levitin F, Stern O, Weiss M, Gil-Henn C, Ziv R, Prokocimer Z, Smorodinsky NI, Rubinstein DB, Wreschner DH. Levitin F, et al. J Biol Chem. 2005 Sep 30;280(39):33374-86. doi: 10.1074/jbc.M506047200. Epub 2005 Jun 29. J Biol Chem. 2005. PMID: 15987679 - Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin.
Macao B, Johansson DG, Hansson GC, Härd T. Macao B, et al. Nat Struct Mol Biol. 2006 Jan;13(1):71-6. doi: 10.1038/nsmb1035. Epub 2005 Dec 20. Nat Struct Mol Biol. 2006. PMID: 16369486 - The role of the SEA (sea urchin sperm protein, enterokinase and agrin) module in cleavage of membrane-tethered mucins.
Palmai-Pallag T, Khodabukus N, Kinarsky L, Leir SH, Sherman S, Hollingsworth MA, Harris A. Palmai-Pallag T, et al. FEBS J. 2005 Jun;272(11):2901-11. doi: 10.1111/j.1742-4658.2005.04711.x. FEBS J. 2005. PMID: 15943821 - Intramembrane proteolysis by presenilin and presenilin-like proteases.
Xia W, Wolfe MS. Xia W, et al. J Cell Sci. 2003 Jul 15;116(Pt 14):2839-44. doi: 10.1242/jcs.00651. J Cell Sci. 2003. PMID: 12808018 Review.
Cited by
- Membrane proximal ectodomain cleavage of MUC16 occurs in the acidifying Golgi/post-Golgi compartments.
Das S, Majhi PD, Al-Mugotir MH, Rachagani S, Sorgen P, Batra SK. Das S, et al. Sci Rep. 2015 Jun 5;5:9759. doi: 10.1038/srep09759. Sci Rep. 2015. PMID: 26044153 Free PMC article. - MUC1 cell surface mucin is a critical element of the mucosal barrier to infection.
McAuley JL, Linden SK, Png CW, King RM, Pennington HL, Gendler SJ, Florin TH, Hill GR, Korolik V, McGuckin MA. McAuley JL, et al. J Clin Invest. 2007 Aug;117(8):2313-24. doi: 10.1172/JCI26705. J Clin Invest. 2007. PMID: 17641781 Free PMC article. - Triple-negative and HER2-overexpressing breast cancer cell sialylation impacts tumor microenvironment T-lymphocyte subset recruitment: a possible mechanism of tumor escape.
Garbar C, Mascaux C, Merrouche Y, Bensussan A. Garbar C, et al. Cancer Manag Res. 2018 May 4;10:1051-1059. doi: 10.2147/CMAR.S162932. eCollection 2018. Cancer Manag Res. 2018. PMID: 29765252 Free PMC article. - Glycocalyx crowding with mucin mimetics strengthens binding of soluble and virus-associated lectins to host cell glycan receptors.
Honigfort DJ, Altman MO, Gagneux P, Godula K. Honigfort DJ, et al. Proc Natl Acad Sci U S A. 2021 Oct 5;118(40):e2107896118. doi: 10.1073/pnas.2107896118. Proc Natl Acad Sci U S A. 2021. PMID: 34583992 Free PMC article. - Membrane-tethered mucins have multiple functions on the ocular surface.
Govindarajan B, Gipson IK. Govindarajan B, et al. Exp Eye Res. 2010 Jun;90(6):655-63. doi: 10.1016/j.exer.2010.02.014. Epub 2010 Mar 16. Exp Eye Res. 2010. PMID: 20223235 Free PMC article. Review.
References
- Abe, J., Suzuki, H., Notoya, M., Yamamoto, T., and Hirose, S. 1999. Ig-hepta, a novel member of the G protein-coupled hepta-helical receptor (GPCR) family that has immunoglobulin-like repeats in a long N-terminal extracellular domain and defines a new subfamily of GPCRs. J. Biol. Chem. 274 19957–19964. - PubMed
- Baruch, A., Hartmann, M., Yoeli, M., Adereth, Y., Greenstein, S., Stadler, Y., Skornik, Y., Zaretsky, J., Smorodinsky, N.I., Keydar, I., and Wreschner, D.H. 1999. The breast cancer-associated MUC1 gene generates both a receptor and its cognate binding protein. Cancer Res. 59 1552–1561. - PubMed
- Cupit, L.D., Schmidt, V.A., and Bahou, W.F. 1999. Proteolytically activated receptor-3. A member of an emerging gene family of protease receptors expressed on vascular endothelial cells and platelets. Trends Cardiovasc. Med. 9 42–48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources