Differential fluorescence induction analysis of Streptococcus pneumoniae identifies genes involved in pathogenesis - PubMed (original) (raw)
Differential fluorescence induction analysis of Streptococcus pneumoniae identifies genes involved in pathogenesis
Andrea Marra et al. Infect Immun. 2002 Mar.
Abstract
Differential fluorescence induction (DFI) technology was used to identify promoters of Streptococcus pneumoniae induced under various in vitro and in vivo conditions. A promoter-trap library using green fluorescent protein as the reporter was constructed in S. pneumoniae, and the entire library was screened for clones exhibiting increased gfp expression under the chosen conditions. The in vitro conditions used were chosen to mimic aspects of the in vivo environment encountered by the pathogen once it enters a host: changes in temperature, osmolarity, oxygen, and iron concentration, as well as blood. In addition, the library was used to infect animals in three different models, and clones induced in these environments were identified. Several promoters were identified in multiple screens, and genes whose promoters were induced twofold or greater under the inducing condition were mutated to assess their roles in virulence. A total of 25 genes were mutated, and the effects of the mutations were assessed in at least two different infection models. Over 50% of these mutants were attenuated in at least one infection model. We show that DFI is a useful tool for identifying bacterial virulence factors as well as a means of elucidating the microenvironment encountered by pathogens upon infection.
Figures
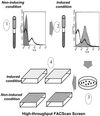
FIG. 1.
High-throughput method of screening individual clones following sorting of the promoter-probe library in S. pneumoniae under a variety of in vitro conditions. When the library is grown under noninducing conditions and analyzed by flow cytometry, the histogram shows peaks of high and low levels of fluorescence (1, heavy line). The shaded histogram represents the vector control strain. When the library is grown under a given inducing condition (see Materials and Methods) and analyzed by flow cytometry, the histogram is shifted, indicating increased fluorescence levels (2, shaded area; the heavy line indicating the uninduced library is shown for comparison). The sorted library is plated for single colonies (3), and individual colonies are picked into wells of microtiter plates for screening under inducing and noninducing conditions (4).
FIG. 2.
Strategy for isolating in vivo-induced promoters. The promoter-probe library (1) in S. pneumoniae is grown to logarithmic phase (2) and used to infect animals in either the respiratory tract, otitis media, or intraperitoneal chamber implant infection model (see Materials and Methods) (3). Bacteria are harvested from the animals and sorted by flow cytometry (4); clones with increased fluorescence are used to reinfect animals for enrichment (5). The sequence of infection and sorting is performed three times, and individual clones harvested from the final round of infection are analyzed as described in Materials and Methods.
FIG. 3.
In vivo characterization of mutants generated after identification in DFI screens. (A) Results of murine respiratory tract infection. The points represent the log number of bacteria harvested from mouse lungs 48 h postinfection. (B) Results of otitis media infection in gerbils. The points represent the log number of bacteria recovered from gerbil middle ear exudates (MEE) 4 days postinfection. (C) Results of otitis media infection in gerbils. The points represent the log number of bacteria found in the blood 4 days following infection. The limit of detection for all experiments is 40 organisms, and the graphs shown are composites of results from at least two individual experiments.
Similar articles
- In vivo characterization of the psa genes from Streptococcus pneumoniae in multiple models of infection.
Marra A, Lawson S, Asundi JS, Brigham D, Hromockyj AE. Marra A, et al. Microbiology (Reading). 2002 May;148(Pt 5):1483-91. doi: 10.1099/00221287-148-5-1483. Microbiology (Reading). 2002. PMID: 11988523 - Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide.
Bartilson M, Marra A, Christine J, Asundi JS, Schneider WP, Hromockyj AE. Bartilson M, et al. Mol Microbiol. 2001 Jan;39(1):126-35. doi: 10.1046/j.1365-2958.2001.02218.x. Mol Microbiol. 2001. PMID: 11123694 - Virulence gene identification by differential fluorescence induction analysis of Staphylococcus aureus gene expression during infection-simulating culture.
Schneider WP, Ho SK, Christine J, Yao M, Marra A, Hromockyj AE. Schneider WP, et al. Infect Immun. 2002 Mar;70(3):1326-33. doi: 10.1128/IAI.70.3.1326-1333.2002. Infect Immun. 2002. PMID: 11854217 Free PMC article. - Role of two-component systems in the virulence of Streptococcus pneumoniae.
Paterson GK, Blue CE, Mitchell TJ. Paterson GK, et al. J Med Microbiol. 2006 Apr;55(Pt 4):355-363. doi: 10.1099/jmm.0.46423-0. J Med Microbiol. 2006. PMID: 16533981 Review. - The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease.
Kadioglu A, Weiser JN, Paton JC, Andrew PW. Kadioglu A, et al. Nat Rev Microbiol. 2008 Apr;6(4):288-301. doi: 10.1038/nrmicro1871. Nat Rev Microbiol. 2008. PMID: 18340341 Review.
Cited by
- Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types.
Pettigrew MM, Fennie KP, York MP, Daniels J, Ghaffar F. Pettigrew MM, et al. Infect Immun. 2006 Jun;74(6):3360-5. doi: 10.1128/IAI.01442-05. Infect Immun. 2006. PMID: 16714565 Free PMC article. - Characterization of central carbon metabolism of Streptococcus pneumoniae by isotopologue profiling.
Härtel T, Eylert E, Schulz C, Petruschka L, Gierok P, Grubmüller S, Lalk M, Eisenreich W, Hammerschmidt S. Härtel T, et al. J Biol Chem. 2012 Feb 3;287(6):4260-74. doi: 10.1074/jbc.M111.304311. Epub 2011 Dec 13. J Biol Chem. 2012. PMID: 22167202 Free PMC article. - Contribution of a response regulator to the virulence of Streptococcus pneumoniae is strain dependent.
Blue CE, Mitchell TJ. Blue CE, et al. Infect Immun. 2003 Aug;71(8):4405-13. doi: 10.1128/IAI.71.8.4405-4413.2003. Infect Immun. 2003. PMID: 12874319 Free PMC article. - Phylogenomic analysis of natural selection pressure in Streptococcus genomes.
Anisimova M, Bielawski J, Dunn K, Yang Z. Anisimova M, et al. BMC Evol Biol. 2007 Aug 30;7:154. doi: 10.1186/1471-2148-7-154. BMC Evol Biol. 2007. PMID: 17760998 Free PMC article.
References
- Badger, J., C. Wass, and K. Kim. 2000. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol. Microbiol. 36:174-182. - PubMed
- Bartilson, M., A. Marra, J. Christine, J. S. Asundi, W. P. Schneider, and A. E. Hromockyj. 2000. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence peptide. Mol. Microbiol. 39:126-135. - PubMed
- Boulnois, G. J. 1992. Pneumococcal proteins and the pathogenesis of disease caused by Streptococcus pneumoniae. J. Gen. Microbiol. 138:249-259. - PubMed
- Canvin, J. R., A. P. Marvin, M. Sivakumaran, J. C. Paton, G. J. Boulnois, P. W. Andrew, and T. J. Mitchell. 1995. The role of pneumolysin and autolysin in the pathology of pneumonia and septicemia in mice infected with a type 2 pneumococcus. J. Infect. Dis. 172:119-123. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical