Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching - PubMed (original) (raw)
Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching
Jesse Stricker et al. Proc Natl Acad Sci U S A. 2002.
Abstract
FtsZ, the major cytoskeletal component of the bacterial cell-division machine, assembles into a ring (the Z-ring) that contracts at septation. FtsZ is a bacterial homolog of tubulin, with similar tertiary structure, GTP hydrolysis, and in vitro assembly. We used green fluorescent protein-labeled FtsZ and fluorescence recovery after photobleaching to show that the E. coli Z-ring is extremely dynamic, continually remodeling itself with a half-time of 30 s. ZipA, a membrane protein involved in cell division that colocalizes with FtsZ, was equally dynamic. The Z-ring of the mutant ftsZ84, which has 1/10 the guanosine triphosphatase activity of wild-type FtsZ in vitro, showed a 9-fold slower turnover in vivo. This finding implies that assembly dynamics are determined primarily by GTP hydrolysis. Despite the greatly reduced assembly dynamics, the ftsZ84 cells divide with a normal cell-cycle time.
Figures
Figure 1
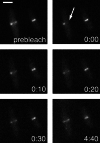
FRAP analysis of FtsZ. MC1000(pBZG) cells were visualized on the microscope slide. Half of the Z-ring of the cell on the left was laser photobleached at the arrow, and recovery of fluorescence was monitored. Panels were photographed at times indicated (minutes:seconds). (Bar = 2 μm.)
Figure 2
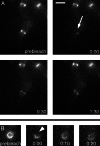
FRAP analysis of the Z-ring in a dividing cell. (Upper Left) Brightfield image of an MC1000(pBZG) cell in the process of division. The cell was photobleached at the arrow and the recovery of fluorescence was monitored. Panels were photographed at times indicated (minutes:seconds). (Bar = 2 μm.)
Figure 3
FRAP analysis of FtsZ84. JFL101(pBZ84G) cells were visualized on the microscope slide. Two cells are outlined in Upper Left. The entire Z-ring of the cell at Upper Right was laser photobleached as indicated by the arrow, and recovery of fluorescence was monitored over time. Panels were photobleached at times indicated (minutes:seconds). (Bar = 2 μm.)
Figure 4
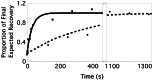
Comparison of recovery of fluorescence of wild-type FtsZ and FtsZ84. Data point values are normalized to the expected final recovery of each species of FtsZ, determined by assuming an exponential recovery with the halftimes given in the text and taking into account the initial loss of fluorophores caused by photobleaching. Note the discontinuous_x_ axis. Filled circles represent time points for the MC1000(pBZG) cell shown in Fig. 1; filled squares represent the JFL101(pBZ84G) cell shown in Fig. 2_A_. The curves indicate theoretical recoveries, given the halftimes of recovery in the text, normalized to final expected recovery as for the experimental data points. The solid curve represents wild-type FtsZ, and the dashed curve represents FtsZ84.
Figure 5
FRAP analysis of ZipA. (A) PB103(λCH50) cells were laser photobleached at the ZipA ring (arrow); recovery of fluorescence was monitored over time. (B) A PB103(λCH50) cell was attached to the coverslip at its end, allowing a face view of the ZipA ring. This cell was photobleached at the arrowhead, and recovery of fluorescence was monitored. Fluorescence recovery appeared to take place at uneven rates around the circumference of this cell. All panels were photographed at times indicated (minutes:seconds). (Bar = 2 μm.)
Figure 6
Two models of how FtsZ protofilaments might be distributed in the Z-ring and cytoplasm. The semicircles are transverse cross sections of 200-nm thickness through the bacterium at the cytoplasm (Left) or Z-ring (Right), and the total number of subunits or protofilaments in this cell volume are shown. (A) A cooperative assembly model in which the Z-ring comprises multifilament bundles, and the cytoplasmic FtsZ is monomeric. In this model, assembly dynamics are thought to involve monomers exchanging onto the ends of the polymers. (B) An isodesmic assembly model. Protofilaments of length 40, 80, and 160 subunits are used to indicate the distribution of lengths around the mean of 80 subunits. In this model, the cytoplasmic FtsZ is largely assembled into protofilaments, and remodeling involves exchanging fragments of protofilaments between the cytoplasm and Z-ring. Attachments of protofilaments to each other and/or to the membrane are not shown but must occur in either model. The models are not exclusive of each other, as assembly could begin with single filaments, which are clustered into multistranded polymers in the Z-ring.
Similar articles
- Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins.
Anderson DE, Gueiros-Filho FJ, Erickson HP. Anderson DE, et al. J Bacteriol. 2004 Sep;186(17):5775-81. doi: 10.1128/JB.186.17.5775-5781.2004. J Bacteriol. 2004. PMID: 15317782 Free PMC article. - ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division.
RayChaudhuri D. RayChaudhuri D. EMBO J. 1999 May 4;18(9):2372-83. doi: 10.1093/emboj/18.9.2372. EMBO J. 1999. PMID: 10228152 Free PMC article. - Mutations in the bacterial cell division protein FtsZ highlight the role of GTP binding and longitudinal subunit interactions in assembly and function.
Arjes HA, Lai B, Emelue E, Steinbach A, Levin PA. Arjes HA, et al. BMC Microbiol. 2015 Oct 13;15:209. doi: 10.1186/s12866-015-0544-z. BMC Microbiol. 2015. PMID: 26463348 Free PMC article. - Bacterial cell division and the septal ring.
Weiss DS. Weiss DS. Mol Microbiol. 2004 Nov;54(3):588-97. doi: 10.1111/j.1365-2958.2004.04283.x. Mol Microbiol. 2004. PMID: 15491352 Review. - Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability.
Romberg L, Levin PA. Romberg L, et al. Annu Rev Microbiol. 2003;57:125-54. doi: 10.1146/annurev.micro.57.012903.074300. Annu Rev Microbiol. 2003. PMID: 14527275 Free PMC article. Review.
Cited by
- 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis.
Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. Strauss MP, et al. PLoS Biol. 2012;10(9):e1001389. doi: 10.1371/journal.pbio.1001389. Epub 2012 Sep 11. PLoS Biol. 2012. PMID: 22984350 Free PMC article. - Lipid Phases and Cell Geometry During the Cell Cycle of Streptococcus pneumoniae.
Calvez P, Jouhet J, Vié V, Durmort C, Zapun A. Calvez P, et al. Front Microbiol. 2019 Mar 18;10:351. doi: 10.3389/fmicb.2019.00351. eCollection 2019. Front Microbiol. 2019. PMID: 30936851 Free PMC article. - Why are bacteria different from eukaryotes?
Theriot JA. Theriot JA. BMC Biol. 2013 Dec 13;11:119. doi: 10.1186/1741-7007-11-119. BMC Biol. 2013. PMID: 24330667 Free PMC article. No abstract available. - ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics.
Camberg JL, Hoskins JR, Wickner S. Camberg JL, et al. Proc Natl Acad Sci U S A. 2009 Jun 30;106(26):10614-9. doi: 10.1073/pnas.0904886106. Epub 2009 Jun 17. Proc Natl Acad Sci U S A. 2009. PMID: 19541655 Free PMC article. - Cysteine 155 plays an important role in the assembly of Mycobacterium tuberculosis FtsZ.
Jaiswal R, Panda D. Jaiswal R, et al. Protein Sci. 2008 May;17(5):846-54. doi: 10.1110/ps.083452008. Protein Sci. 2008. PMID: 18436955 Free PMC article.
References
- Bramhill D. Annu Rev Cell Dev Biol. 1997;13:395–424. - PubMed
- Lutkenhaus J, Addinall S G. Annu Rev Biochem. 1997;66:93–116. - PubMed
- Rothfield L, Justice S, Garcia-Lara J. Annu Rev Genet. 1999;33:423–448. - PubMed
- Löwe J. J Struct Biol. 1998;124:235–243. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources