Budding of equine infectious anemia virus is insensitive to proteasome inhibitors - PubMed (original) (raw)
Budding of equine infectious anemia virus is insensitive to proteasome inhibitors
Akash Patnaik et al. J Virol. 2002 Mar.
Abstract
The only retrovirus protein required for the budding of virus-like particles is the Gag protein; however, recent studies of Rous sarcoma virus (RSV) and human immunodeficiency virus have suggested that modification of Gag with ubiquitin (Ub) is also required. As a consequence, the release of these viruses is reduced in the presence of proteasome inhibitors, which indirectly reduce the levels of free Ub within the cell. Here we show that the budding of equine infectious anemia virus (EIAV) from infected equine cells is largely unaffected by these drugs, although use of one inhibitor (MG-132) resulted in a dramatic block to proteolytic processing of Gag. This lack of sensitivity was also observed in transiently transfected avian cells under conditions that greatly reduce RSV budding. Moreover, insensitivity was observed when the EIAV Gag protein was expressed in the absence of all the other virus products, indicating that they are not required for this phenotype. An activity that enables EIAV to tolerate exposure to proteasome inhibitors was mapped to the C-terminal p9 sequence, as demonstrated by the ability of an RSV Gag-p9 chimera to bud in the presence of the drugs. Intriguingly, the p9 sequence contains a short sequence motif that is similar to a surface-exposed helix of Ub, suggesting that EIAV Gag may have captured a function that allows it to bypass the need for ubiquitination. Thus, the mechanism of EIAV budding may not be substantially different from that of other retroviruses, even though it behaves differently in the presence of proteasome inhibitors.
Figures
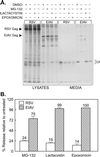
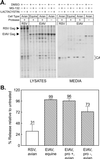
FIG. 1.
Insensitivity of EIAV release to proteasome inhibitors. (A) Four plates each of RSV-infected avian cells and EIAV-infected equine cells were seeded, and, the next day, one from each set was pretreated for 90 min with DMSO (vehicle control), 10 μM MG-132, 10 μM lactacystin, or 10 μM epoxomicin. The cells were then metabolically labeled for 2.5 h with [35S]methionine, also in the presence of drugs. The cell monolayers and the particles in the growth medium were lysed with detergent, and the Gag proteins were immunoprecipitated and electrophoresed in an SDS-12% polyacrylamide gel prior to detection by autoradiography. Left, positions of the uncleaved Gag proteins; right, positions of the mature CA products. The majority of methionine residues in EIAV Gag (10 of 12) are found in CA, and hence it is the only cleavage product visible. (B) The impact of proteasome inhibitors on RSV and EIAV budding was quantitated with a phosphorimager. The budding efficiency in each culture was calculated as the total amount of Gag protein (Gag precursor plus intermediate cleavage products plus mature cleavage products) in the medium divided by the total amount in the culture (lysates and medium). The effects of proteasome inhibitors on budding (percent release relative to untreated) were then determined by computing the ratio of budding efficiency in the DMSO-treated cells to that in drug-treated cells. Averages from two experiments are shown, with error bars indicating the deviation of each measurement from the mean.
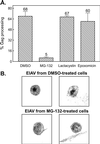
FIG. 2.
Inhibitory effects of MG-132 on EIAV protease activity. (A) EIAV-infected cells were labeled with [35S]methionine as indicated for Fig. 1. The impact of the proteasome inhibitor on protease activity is expressed as the ratio of mature cleavage products (CA) in the medium to the total Gag signal (Gag precursor plus intermediate cleavage products plus mature cleavage products) in the medium. Averages from two experiments are shown along with the deviation of each measurement from the mean. (B) Transmission electron microscopy of the surfaces of EIAV-infected equine cells treated with either DMSO (top) or 80 μM MG-132 (bottom).
FIG. 3.
Direct comparison of EIAV and RSV budding in avian cells. (A) Triplicate plates of uninfected cells were transfected with CMV promoter-driven expression vectors containing the wild-type EIAV structural genes (pCMV.EIAVuk) or a protease mutant (pCMV.EIAVuk.pro-). In addition, triplicate plates of RSV-infected avian cells (positive control) and EIAV-infected equine cells (negative control) were seeded. The next day, plates from each set were pretreated for 90 min with DMSO (control), MG-132, or lactacystin, and the cells were metabolically labeled for 2.5 h with [35S]methionine, also in the presence of the drug. The cells and growth media were subsequently processed as described for Fig. 1. (B) The impact of lactacystin on budding was quantitated by phosphorimager, as described for Fig. 1. Averages from two experiments are shown, with error bars indicating the deviations of each measurement from the mean.
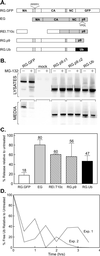
FIG. 4.
Drug resistance activity maps to a region within p9Gag. (A) Gag derivatives used for the mapping experiments. Open boxes, segments of the RSV Gag protein (along with GFP); gray boxes, segments of EIAV Gag. The relative positions of the RSV and EIAV late domains are indicated. (B) QT6 cells were transfected with the indicated DNAs. The next day, the cells were pretreated with either DMSO or 80 μM MG-132 for 90 min, before being metabolically labeled for 2.5 h with [35S]Met, also in the presence of the drug. The cells and growth media were subsequently processed as described for Fig. 1. (C) Phosphorimager analysis of budding efficiencies, calculated as described for Fig. 1. Averages from three experiments are shown for all constructs, with the exception of REI.T10c, which shows data from two experiments. (D) Reduction in free-Ub levels with time following exposure to 80 μM MG-132. Avian cell lysates were prepared at the indicated times and analyzed on an immunoblot using antibodies specific for Ub. Bands on the resulting autoradiograph were quantitated by means of densitometry.
FIG. 5.
A ubiquitin-like motif within EIAV p9Gag. Alignment of EIAV p9Gag and ubiquitin showing the region of similarity (black rectangles) between the two proteins. The position of the L domain is also indicated (gray rectangle).
Similar articles
- Equine infectious anemia virus and the ubiquitin-proteasome system.
Ott DE, Coren LV, Sowder RC 2nd, Adams J, Nagashima K, Schubert U. Ott DE, et al. J Virol. 2002 Mar;76(6):3038-44. doi: 10.1128/jvi.76.6.3038-3044.2002. J Virol. 2002. PMID: 11861870 Free PMC article. - Late domain-dependent inhibition of equine infectious anemia virus budding.
Shehu-Xhilaga M, Ablan S, Demirov DG, Chen C, Montelaro RC, Freed EO. Shehu-Xhilaga M, et al. J Virol. 2004 Jan;78(2):724-32. doi: 10.1128/jvi.78.2.724-732.2004. J Virol. 2004. PMID: 14694104 Free PMC article. - Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection.
Chen C, Li F, Montelaro RC. Chen C, et al. J Virol. 2001 Oct;75(20):9762-70. doi: 10.1128/JVI.75.20.9762-9770.2001. J Virol. 2001. PMID: 11559809 Free PMC article. - Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein.
Puffer BA, Parent LJ, Wills JW, Montelaro RC. Puffer BA, et al. J Virol. 1997 Sep;71(9):6541-6. doi: 10.1128/JVI.71.9.6541-6546.1997. J Virol. 1997. PMID: 9261374 Free PMC article. - Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication.
Li F, Chen C, Puffer BA, Montelaro RC. Li F, et al. J Virol. 2002 Feb;76(4):1569-77. doi: 10.1128/jvi.76.4.1569-1577.2002. J Virol. 2002. PMID: 11799151 Free PMC article.
Cited by
- Role of ESCRT-I in retroviral budding.
Martin-Serrano J, Zang T, Bieniasz PD. Martin-Serrano J, et al. J Virol. 2003 Apr;77(8):4794-804. doi: 10.1128/jvi.77.8.4794-4804.2003. J Virol. 2003. PMID: 12663786 Free PMC article. - Identification of conserved motifs in the West Nile virus envelope essential for particle secretion.
Garg H, Lee RT, Tek NO, Maurer-Stroh S, Joshi A. Garg H, et al. BMC Microbiol. 2013 Sep 4;13:197. doi: 10.1186/1471-2180-13-197. BMC Microbiol. 2013. PMID: 24007503 Free PMC article. - Equine viperin restricts equine infectious anemia virus replication by inhibiting the production and/or release of viral Gag, Env, and receptor via distortion of the endoplasmic reticulum.
Tang YD, Na L, Zhu CH, Shen N, Yang F, Fu XQ, Wang YH, Fu LH, Wang JY, Lin YZ, Wang XF, Wang X, Zhou JH, Li CY. Tang YD, et al. J Virol. 2014 Nov;88(21):12296-310. doi: 10.1128/JVI.01379-14. Epub 2014 Aug 13. J Virol. 2014. PMID: 25122784 Free PMC article. - Analysis of human immunodeficiency virus type 1 Gag ubiquitination.
Gottwein E, Kräusslich HG. Gottwein E, et al. J Virol. 2005 Jul;79(14):9134-44. doi: 10.1128/JVI.79.14.9134-9144.2005. J Virol. 2005. PMID: 15994808 Free PMC article. - Antiviral activity of α-helical stapled peptides designed from the HIV-1 capsid dimerization domain.
Zhang H, Curreli F, Zhang X, Bhattacharya S, Waheed AA, Cooper A, Cowburn D, Freed EO, Debnath AK. Zhang H, et al. Retrovirology. 2011 May 3;8:28. doi: 10.1186/1742-4690-8-28. Retrovirology. 2011. PMID: 21539734 Free PMC article.
References
- Cunningham, T. P., R. C. Montelaro, and K. E. Rushlow. 1993. Lentivirus envelope sequences and proviral genomes are stabilized in Escherichia coli when cloned in low-copy-number plasmid vectors. Gene 124:93-98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA047482/CA/NCI NIH HHS/United States
- R01 CA049296/CA/NCI NIH HHS/United States
- CA47484/CA/NCI NIH HHS/United States
- CA49296/CA/NCI NIH HHS/United States
- R37 CA047482/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials