Exploring prokaryotic diversity in the genomic era - PubMed (original) (raw)
Review
Exploring prokaryotic diversity in the genomic era
Philip Hugenholtz. Genome Biol. 2002.
Abstract
Our understanding of prokaryote biology from study of pure cultures and genome sequencing has been limited by a pronounced sampling bias towards four bacterial phyla - Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes - out of 35 bacterial and 18 archaeal phylum-level lineages. This bias is beginning to be rectified by the use of phylogenetically directed isolation strategies and by directly accessing microbial genomes from environmental samples.
Figures
Figure 1
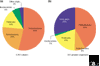
Pie charts showing the phylum-level distribution of prokaryotic isolates (a) in the Australian Collection of Microorganisms [11] and (b) in the prokaryote genome sequences completed or in progress as of 20 August 2001 [29].
Figure 2
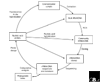
'Full-cycle' rRNA approach to characterizing microorganisms in their natural settings without the need for cultivation. Access to whole genomes of uncultivated organisms is also possible using the same basic approach but with large-insert cloning vectors, such as BACs, which remove the need for PCR.
Figure 3
Evolutionary distance dendrograms of (a) bacterial and (b) archaeal diversity derived from comparative analysis of 16S rRNA gene sequences. The trees were constructed using the ARBsoftware package and a sequence database modified from the March 1997 ARB database release [39] using 50% consensus sequence filters for each domain and the Olsen correction and neighbor-joining options. This modified database will be available from the Ribosomal Database Project [40] user-submitted alignments download site [41]. Major lineages (phyla) are shown as wedges with horizontal dimensions reflecting the known degree of divergence within that lineage. Phyla with cultivated representatives are in gray and, where possible, named according to the taxonomic outline of Bergey's Manual [8]. Phyla known only from environmental sequences are in white; because they are not formally recognized as taxonomic groups, they are usually named after the first clones found from within the group [14,20]. Note that environmental groups E2 and E3 defined in [20] are part of the Thermoplasmata phylum in the archaeal tree in (b). The number of genome sequences completed or in progress for each phylum is given in brackets after the phylum name, with the exception of Methanopyrus kandleri, which is not included in the tree because it is represented by a single sequence. The scale bar represents 0.1 changes per nucleotide.
Similar articles
- Exploring cultivable Bacteria from the prokaryotic community associated with the carnivorous sponge Asbestopluma hypogea.
Dupont S, Carre-Mlouka A, Domart-Coulon I, Vacelet J, Bourguet-Kondracki ML. Dupont S, et al. FEMS Microbiol Ecol. 2014 Apr;88(1):160-74. doi: 10.1111/1574-6941.12279. Epub 2014 Feb 10. FEMS Microbiol Ecol. 2014. PMID: 24392789 - Proteobacteria and Bacteroidetes are major phyla of filterable bacteria passing through 0.22 μm pore size membrane filter, in Lake Sanaru, Hamamatsu, Japan.
Maejima Y, Kushimoto K, Muraguchi Y, Fukuda K, Miura T, Yamazoe A, Kimbara K, Shintani M. Maejima Y, et al. Biosci Biotechnol Biochem. 2018 Jul;82(7):1260-1263. doi: 10.1080/09168451.2018.1456317. Epub 2018 Mar 29. Biosci Biotechnol Biochem. 2018. PMID: 29598452 - Eubacterial diversity of activated biomass from a common effluent treatment plant.
Kapley A, De Baere T, Purohit HJ. Kapley A, et al. Res Microbiol. 2007 Jul-Aug;158(6):494-500. doi: 10.1016/j.resmic.2007.04.004. Epub 2007 Apr 27. Res Microbiol. 2007. PMID: 17566710 - Evolution of transcriptional regulation of histidine metabolism in Gram-positive bacteria.
Ashniev GA, Sernova NV, Shevkoplias AE, Rodionov ID, Rodionova IA, Vitreschak AG, Gelfand MS, Rodionov DA. Ashniev GA, et al. BMC Genomics. 2022 Aug 25;23(Suppl 6):558. doi: 10.1186/s12864-022-08796-y. BMC Genomics. 2022. PMID: 36008760 Free PMC article. - Molecular phylogeny of Eubacteria: a new multiple tree analysis method applied to 15 sequence data sets questions the monophyly of gram-positive bacteria.
Galtier N, Gouy M. Galtier N, et al. Res Microbiol. 1994 Sep;145(7):531-41. doi: 10.1016/0923-2508(94)90030-2. Res Microbiol. 1994. PMID: 7855439 Review.
Cited by
- Most of the Dominant Members of Amphibian Skin Bacterial Communities Can Be Readily Cultured.
Walke JB, Becker MH, Hughey MC, Swartwout MC, Jensen RV, Belden LK. Walke JB, et al. Appl Environ Microbiol. 2015 Oct;81(19):6589-600. doi: 10.1128/AEM.01486-15. Epub 2015 Jul 10. Appl Environ Microbiol. 2015. PMID: 26162880 Free PMC article. - Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics.
Alcaraz LD, Moreno-Hagelsieb G, Eguiarte LE, Souza V, Herrera-Estrella L, Olmedo G. Alcaraz LD, et al. BMC Genomics. 2010 May 26;11:332. doi: 10.1186/1471-2164-11-332. BMC Genomics. 2010. PMID: 20504335 Free PMC article. - Novel microarray design strategy to study complex bacterial communities.
Huyghe A, Francois P, Charbonnier Y, Tangomo-Bento M, Bonetti EJ, Paster BJ, Bolivar I, Baratti-Mayer D, Pittet D, Schrenzel J; Geneva Study Group on Noma (GESNOMA). Huyghe A, et al. Appl Environ Microbiol. 2008 Mar;74(6):1876-85. doi: 10.1128/AEM.01722-07. Epub 2008 Jan 18. Appl Environ Microbiol. 2008. PMID: 18203854 Free PMC article. - A metagenomic study of methanotrophic microorganisms in Coal Oil Point seep sediments.
Håvelsrud OE, Haverkamp TH, Kristensen T, Jakobsen KS, Rike AG. Håvelsrud OE, et al. BMC Microbiol. 2011 Oct 4;11:221. doi: 10.1186/1471-2180-11-221. BMC Microbiol. 2011. PMID: 21970369 Free PMC article. - The case for biocentric microbiology.
Aziz RK. Aziz RK. Gut Pathog. 2009 Aug 4;1(1):16. doi: 10.1186/1757-4749-1-16. Gut Pathog. 2009. PMID: 19653908 Free PMC article.
References
- Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. - PubMed
- Galvez A, Maqueda M, Martinez-Bueno M, Valdivia E. Publication rates reveal trends in microbiological research. ASM News. 1998;64:269–275.
- DSMZ Bacterial Nomenclature Up-to-date http://www.dsmz.de/bactnom/bactname.htm
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources