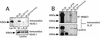Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors - PubMed (original) (raw)
Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors
Ari Melnick et al. Mol Cell Biol. 2002 Mar.
Abstract
The PLZF (promyelocytic leukemia zinc finger) transcriptional repressor, when fused to retinoic acid receptor alpha (RARalpha), causes a refractory form of acute promyelocytic leukemia. The highly conserved N-terminal BTB (bric a brac, tramtrack, broad complex)/POZ domain of PLZF plays a critical role in this disease, since it is required for transcriptional repression by the PLZF-RARalpha fusion protein. The crystal structure of the PLZF BTB domain revealed an obligate homodimer with a highly conserved charged pocket formed by apposition of the two monomers. An extensive structure-function analysis showed that the charged pocket motif plays a major role in transcriptional repression by PLZF. We found that mutations of the BTB domain that neutralize key charged pocket residues did not disrupt dimerization, yet abrogated the ability of PLZF to repress transcription and led to the loss of interaction with N-CoR, SMRT, and histone deacetylases (HDACs). We extended these studies to the Bcl-6 protein, which is linked to the pathogenesis of non-Hodgkin's lymphomas. In this case, neutralizing the charged pocket also resulted in loss of repression and corepressor binding. Experiments with purified protein showed that corepressor-BTB interactions were direct. A comparison of the PLZF, Bcl-6, and the FAZF (Fanconi anemia zinc finger)/ROG protein shows that variations in the BTB pocket result in differential affinity for corepressors, which predicts the potency of transcriptional repression. Thus, the BTB pocket represents a molecular structure involved in recruitment of transcriptional repression complexes to target promoters.
Figures
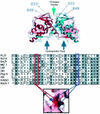
FIG. 1.
Structural motifs of the BTB dimer. (Top panel) Superimposed ribbon and charge-mapping views of the PLZF BTB dimer (red, negative charge; blue, positive charge). The BTB monomer ribbons are shown in blue and red, respectively. The green arrow indicates the highly conserved charged pocket, and the blue arrows indicate the hydrophobic oligomerization surface region. The thin black arrows indicate the location of key, conserved charged pocket residues in both monomers. (Middle panel) Sequence alignment of the N terminus of several different BTB domains. This region of the BTB corresponds mainly to the charged pocket. The red box indicates the conserved aspartate residue, which in PLZF is at position 35. The blue box indicates the conserved basic pocket residue, which in PLZF is an arginine at position 49. The location of these two residues in the charged pocket is shown in the lower panel, which is a charge mapping viewed from the “top” of the dimer.
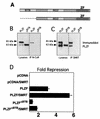
FIG. 2.
The BTB domain is physically and functionally critical for interactions with corepressors. (A) PLZF contains discrete functional domains, including the N-terminal BTB domain, a less-well-characterized second repression domain (RD2), and nine C-terminal Zn fingers (ZF). The PLZFΔBTB construct used in these experiments is also shown. (B) Coimmunoprecipitations (IP) performed in 293T cells. Cells (4 × 105) were transfected with 1 μg of PLZF or PLZFΔBTB expression vectors. Endogenous N-CoR was precipitated, and the resulting fraction was immunoblotted to detect PLZF. Expression of both proteins is confirmed by direct Western blotting of the cell lysates as shown in the left two lanes. The lower band in the PLZF lanes is an alternate PLZF product commonly seen in cells transfected with the PLZF expression vector. (C) Coimmunoprecipitations with endogenous SMRT similar to panel B. (D) 293T cells (2 × 105) were transfected with 200 ng of pCDNA, pCDNA-PLZF, or pCDNA-PLZFΔBTB, with or without 150 ng of CMX-SMRT. The cells were cotransfected with 50 ng of a luciferase reporter containing four PLZF binding sites from the IL-3Rα chain promoter and a _tk_-renilla internal control. Results are expressed as fold repression of luciferase normalized to the internal control.
FIG. 3.
The PLZF BTB charged pocket is required for transcriptional repression and corepressor interaction. (A) 293T cells (2 × 105) were cotransfected with 200 ng of the indicated GAL-fusion effectors, 50 ng of (GAL)5-_tk_-Luc reporter, and 5 ng of _tk_-renilla. Results are expressed as fold repression of luciferase normalized to the internal control. (B) 293T cells (2 × 105) were cotransfected with 200 ng of the indicated wild-type and mutant PLZF species, 50 ng of (IL-3Rα)4-_tk_-Luc reporter, and 5 ng of _tk_-renilla. (C) The ability of N-CoR to precipitate BTB domains was tested by allowing in vitro-translated N-CoR to interact with in vitro-translated BTB domains. Both proteins were 35S labeled and visualized by fluorography. The top gel shows BTB domains after precipitation with N-CoR antibodies; the middle and bottom gels show the presence of the indicated BTB species N-CoR in the input lysates. IP, immunoprecipitation. (D) Western blots of transfected full-length PLZF proteins with wild-type or point mutant BTB domains after immunoprecipitation with N-CoR antibodies. The lysate input and corresponding immunoprecipitation reactions are paired in each of the boxes shown. (E) PLZF monoclonal antibody was used to immunoprecipitate lysates from 293T cells transfected with full-length wild-type PLZF and PLZF harboring mutations in the BTB domain. Western blots were performed with the resolved proteins to determine the presence of the SMRT corepressor. Lane 1 shows the presence of SMRT in the cell lysates. (F) Similar to panel A, 293T cells were cotransfected with GAL-BTB fusions and the GAL reporter, with or without 150 ng of SMRT, as indicated. (G) 293T cells were transfected as shown in panel B with full-length PLZF or the indicated mutants along with the IL-3Rα reporter, with or without 150 ng of SMRT.
FIG. 4.
The PLZF BTB charged pocket is required for efficient recruitment of HDACs. 293T cells (106) were transfected with 1 μg of full-length PLZF or mutants and 1 μg of HDAC1 expression plasmid. (A) Lysates were subjected to immunoprecipitation (IP) with PLZF antibodies and then immunoblotted with HDAC1 antibodies. The first lane shows lysate input (Lys), followed by lysates after precipitation with preimmune rabbit sera and then respective coprecipitations with wild-type and mutant forms of PLZF. The lower gel shows expression of HDAC- 1 in the input lysates. (B) Precipitations were performed with HDAC1-specific antibodies, and the resulting proteins were immunoblotted with PLZF antisera. The PLZF species used in each case is indicated. Lysate controls are shown below.
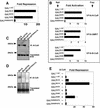
FIG. 5.
The charged pocket is required for Bcl-6 BTB transcriptional repression and interaction with corepressors. Wild-type and mutant Bcl-6 BTB domains were analyzed in several assays. (A) 293T cells (2 × 105) were transfected with 100 ng of GAL-Bcl-6 expression vectors and 50 ng of the (GAL)5-_tk_-Luc reporter and 5 ng of _tk_-renilla as an internal control. The results are expressed as fold repression of luciferase normalized to the internal control. (B) Mammalian two-hybrid assays performed in 293T cells transfected with either 25 ng of GAL-Bcl-6BTB bait plasmids and 150 ng of VP16-N-CoR or -SMRT prey. HeLa cells were transfected with 5 ng of GAL-Bcl-6BTB bait and 25 ng of VP16-B-CoR prey. Results are expressed as fold activation of the (GAL)5-_tk_-Luc reporter normalized to the internal control (_tk_-renilla in N-CoR and SMRT experiments and a cytomegalovirus-β-galactosidase [CMV-β-Gal] construct in B-CoR experiments in HeLa cells). B-CoR interaction was also tested in 293T cells with similar results (not shown). (C) In vitro-translated 35S-labeled Bcl-6 wild-type and mutant BTB domains were mixed with in vitro-translated N-CoR. Lysates were precipitated with N-CoR antibodies and resolved by fluorography. The top gel shows the presence of BTB domains after coimmunoprecipitation (IP), and the bottom gel verifies the presence of the BTB domains in the lysates prior to precipitation. (D) In vitro-translated Bcl-6 BTB species were allowed to interact with myc-tagged B-CoR and immunoprecipitated with myc epitope antibodies. The top gel shows lysates after coimmunoprecipitation, and the bottom gel shows the lysates prior to precipitation. (E) Reporter assays were performed as in panel A. The ability of B-CoR to enhance repression by the wild type or mutant Bcl-6BTB was determined by transfecting 100 ng of the B-CoR expression plasmid to the indicated cell cultures.
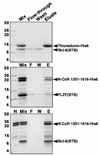
FIG. 6.
A discrete domain of N-CoR and SMRT interacts directly with the BTB domains of PLZF and Bcl-6. In vitro interaction of Bcl-6BTB and PLZFBTB with a fragment of N-CoR. Purified BTB domain was mixed in solution with the indicated histidine-tagged protein, followed by Ni-NTA affinity purification. The samples were resolved on SDS-PAGE gel (14% polyacrylamide) followed by Coomassie staining. Molecular weight markers are indicated. Lanes: Mix, mixture before loading on the Ni-NTA column; F, column flowthrough (unbound protein); W, column wash; E, imidazole eluate (releases His-tagged proteins); N (bottom gel), purified residues 1351 to 1616 before mixing with BTB domain. The N-CoR fragment is a doublet; the smaller band corresponds to a partial proteolysis product that retains the His tag. Bcl-6BTB interacts strongly with the N-CoR fragment, while the PLZFBTB domains interact, but to a lesser extent. Neither interacts with His-tagged thioredoxin.
FIG. 7.
Transcriptional repression and corepressor binding correlate with the identity of the critical charged pocket residues in PLZF, Bcl-6, and FAZF. (A) Sequence alignment of the pocket region of the BTB domains of PLZF, Bcl-6, and FAZF. The arrowheads indicate the critical charged residues: D35 in PLZF (this is D33 in Bcl-6 and D29 in FAZF) and R49 in PLZF (this is K47 in Bcl-6 and S44 in FAZF). The S44 residue of FAZF is circled. (B) 293T cells were transfected with 400 ng of the GAL1-147 or respective GAL-BTB fusions from PLZF, Bcl-6, or FAZF along with 100 ng of (GAL)5-_tk_-Luc reporter and 5 ng of _tk_-renilla control plasmids. (C) Mammalian two-hybrid experiments were performed with 25 ng of the indicated GAL constructs as bait and 150 ng of VP16-N-CoR or -SMRT as prey. The results are expressed as fold activation of the luciferase reporter normalized to the internal control. (D) GAL4 DBD immunoblots corresponding to the mammalian two-hybrid assay described in panel C were performed in 293T cells transfected with equivalent amounts of GAL-BTB fusions and GAL-VP16 prey plasmids as indicated. Lanes: N, VP16-N-CoR cotransfection; lane S: VP16-SMRT cotransfection.
FIG. 8.
Swapping critical Bcl-6 charged pocket residues alters transcriptional repression and corepressor binding properties. Mutant Bcl-6 BTB domains were generated by swapping the K47 residue for arginine (as in PLZF) or serine (as in FAZF). (A) Reporter assays in 293T cells transfected with 25 ng of GAL1-147 or GAL-Bcl-6BTB mutants as indicated along with 100 ng of reporter and 5 ng of the internal control. Results are tabulated as fold repression compared to that of GAL1-147 normalized to the internal control. (B) Mammalian two-hybrid assays in 293T cells transfected with 25 ng of GAL-Bcl-6 bait and 150 ng of VP-16-N-CoR or -SMRT prey or 5 ng of GAL-Bcl-6 bait with 25 ng of VP16-B-CoR prey in HeLa cells. Results are reported as fold activation of the (GAL)5-_tk_-Luc reporter construct. B-CoR interaction was also tested in 293 T cells with similar results (not shown). (C) Coimmunoprecipitations (IP) were performed with in vitro-translated BTB domains and N-CoR as described previously. The top section shows fluorography of BTB domains precipitated with N-CoR antibodies, and the bottom section shows the appropriate lysates prior to immunoprecipitation.
FIG. 9.
Swapping critical charged pocket residues in FAZF and PLZF alters transcriptional repression and corepressor interactions. Mutant PLZF BTB domains were generated by swapping the R49 residue for lysine (as in Bcl-6) or serine (as in FAZF). FAZF mutants were also made by swapping the S44 for lysine (as in Bcl-6) or arginine (as in PLZF). (A and B) Reporter assays performed with 2 × 105 293T cells cotransfected with 400 ng of GAL1-147 or GAL-BTB fusions as indicated with 100 ng of reporter and 5 ng of internal control. (C) Mammalian two-hybrid assay performed in 293T cells with 25 ng of GAL1-147 or GAL-PLZFBTB bait and 150 ng of VP16-SMRT prey as indicated.
Similar articles
- In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions.
Melnick A, Ahmad KF, Arai S, Polinger A, Ball H, Borden KL, Carlile GW, Prive GG, Licht JD. Melnick A, et al. Mol Cell Biol. 2000 Sep;20(17):6550-67. doi: 10.1128/MCB.20.17.6550-6567.2000. Mol Cell Biol. 2000. PMID: 10938130 Free PMC article. - Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: the case of HIC-1 and gammaFBP-B.
Deltour S, Guerardel C, Leprince D. Deltour S, et al. Proc Natl Acad Sci U S A. 1999 Dec 21;96(26):14831-6. doi: 10.1073/pnas.96.26.14831. Proc Natl Acad Sci U S A. 1999. PMID: 10611298 Free PMC article. - The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT.
Huynh KD, Bardwell VJ. Huynh KD, et al. Oncogene. 1998 Nov 12;17(19):2473-84. doi: 10.1038/sj.onc.1202197. Oncogene. 1998. PMID: 9824158 - N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors.
Jones PL, Shi YB. Jones PL, et al. Curr Top Microbiol Immunol. 2003;274:237-68. doi: 10.1007/978-3-642-55747-7_9. Curr Top Microbiol Immunol. 2003. PMID: 12596910 Review. - Biological roles and mechanistic actions of co-repressor complexes.
Jepsen K, Rosenfeld MG. Jepsen K, et al. J Cell Sci. 2002 Feb 15;115(Pt 4):689-98. doi: 10.1242/jcs.115.4.689. J Cell Sci. 2002. PMID: 11865025 Review.
Cited by
- B Cell Lymphoma 6 (BCL6): A Conserved Regulator of Immunity and Beyond.
Liongue C, Almohaisen FLJ, Ward AC. Liongue C, et al. Int J Mol Sci. 2024 Oct 11;25(20):10968. doi: 10.3390/ijms252010968. Int J Mol Sci. 2024. PMID: 39456751 Free PMC article. Review. - Critical domains for NACC2-NTRK2 fusion protein activation.
Yang W, Meyer AN, Jiang Z, Jiang X, Donoghue DJ. Yang W, et al. PLoS One. 2024 Jun 27;19(6):e0301730. doi: 10.1371/journal.pone.0301730. eCollection 2024. PLoS One. 2024. PMID: 38935636 Free PMC article. - Distinct Perception Mechanisms of BACH1 Quaternary Structure Degrons by Two F-box Proteins under Oxidative Stress.
Cao S, Shi H, Garcia SF, Kito Y, Shi H, Goldberg HV, Ponce J, Ueberheide B, Lignitto L, Pagano M, Zheng N. Cao S, et al. bioRxiv [Preprint]. 2024 Jun 3:2024.06.03.594717. doi: 10.1101/2024.06.03.594717. bioRxiv. 2024. PMID: 38895309 Free PMC article. Updated. Preprint. - Znf687 recruits Brd4-Smrt complex to regulate gfi1aa during neutrophil development.
Yan L, Tan S, Wang H, Yuan H, Liu X, Chen Y, de Thé H, Zhu J, Zhou J. Yan L, et al. Leukemia. 2024 Apr;38(4):851-864. doi: 10.1038/s41375-024-02165-2. Epub 2024 Feb 7. Leukemia. 2024. PMID: 38326409 - The N-Terminal Part of Drosophila CP190 Is a Platform for Interaction with Multiple Architectural Proteins.
Golovnin A, Melnikova L, Babosha V, Pokholkova GV, Slovohotov I, Umnova A, Maksimenko O, Zhimulev IF, Georgiev P. Golovnin A, et al. Int J Mol Sci. 2023 Nov 2;24(21):15917. doi: 10.3390/ijms242115917. Int J Mol Sci. 2023. PMID: 37958900 Free PMC article.
References
- Barna, M., N. Hawe, L. Niswander, and P. P. Pandolfi. 2000. Plzf regulates limb and axial skeletal patterning. Nat. Genet. 25:166-172. - PubMed
- Cattoretti, G., C. C. Chang, K. Cechova, J. Zhang, B. H. Ye, B. Falini, D. C. Louie, K. Offit, R. S. Chaganti, and R. Dalla-Favera. 1995. BCL-6 protein is expressed in germinal-center B cells. Blood 86:45-53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA059936/CA/NCI NIH HHS/United States
- K08 CA 73762/CA/NCI NIH HHS/United States
- R29 CA071540/CA/NCI NIH HHS/United States
- R01 CA 59936/CA/NCI NIH HHS/United States
- R29 CA 71540/CA/NCI NIH HHS/United States
- R01 CA071540/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous