Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing - PubMed (original) (raw)
Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing
Gunnar Schotta et al. EMBO J. 2002.
Abstract
Su(var)3-9 is a dominant modifier of heterochromatin-induced gene silencing. Like its mammalian and Schizosaccharomyces pombe homologues, Su(var) 3-9 encodes a histone methyltransferase (HMTase), which selectively methylates histone H3 at lysine 9 (H3-K9). In Su(var)3-9 null mutants, H3-K9 methylation at chromocentre heterochromatin is strongly reduced, indicating that SU(VAR)3-9 is the major heterochromatin-specific HMTase in Drosophila. SU (VAR)3-9 interacts with the heterochromatin-associated HP1 protein and with another silencing factor, SU(VAR)3-7. Notably, SU(VAR)3-9-HP1 interaction is interdependent and governs distinct localization patterns of both proteins. In Su(var)3-9 null mutants, concentration of HP1 at the chromocentre is nearly lost without affecting HP1 accumulation at the fourth chromosome. By contrast, in HP1 null mutants SU(VAR)3-9 is no longer restricted at heterochromatin but broadly dispersed across the chromosomes. Despite this interdependence, Su(var)3-9 dominates the PEV modifier effects of HP1 and Su(var)3-7 and is also epistatic to the Y chromosome effect on PEV. Finally, the human SUV39H1 gene is able to partially rescue Su(var)3-9 silencing defects. Together, these data indicate a central role for the SU(VAR)3-9 HMTase in heterochromatin-induced gene silencing in Drosophila.
Figures
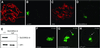
Fig. 1. Distribution of the SU(VAR)3–9 protein in salivary gland polytene chromosomes. (A and B) Immunostaining of polytene chromosomes from a pP{GS[ry+, hs (Su(var)3–9 cDNA)–EGFP]} transgenic line with a rabbit polyclonal α-GFP antibody. (C and D) Heterochromatin association of endogenous SU(VAR)3–9 after staining with a rabbit polyclonal α-SU(VAR)3–9 antibody. DNA staining with propidium iodide (A and C) and immunostaining of the same nuclei, respectively (B and D). (E) Western blot of nuclear extracts from wild-type and Su(var)3–9 null mutant embryos. Blot probed with α-SU(VAR)3–9 antibody (upper panel) and α-HP1 antibody (lower panel). (F–H) Confocal sections of salivary gland nuclei showing in vivo distribution of SU(VAR)3–9–EGFP in wild-type (F), T(2;3)TE80var20/+; pP{GS[ry+, hs (Su(var)3–9 cDNA)–EGFP]} (G) and _w_m4/_w_m4; pP{GS[ry+, hs (Su(var)3–9 cDNA)–EGFP]} (H) larvae. The chromosomal rearrangements relocate regions of heterochromatin to distal euchromatin (arrows).
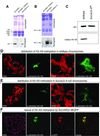
Fig. 2. Histone H3-K9 methylation in heterochromatin is controlled by SU(VAR)3–9. (A and B) H3-K9 HMTase activity of recombinant SU(VAR)3–9 protein for histone H3 (A) and N-terminal peptide (1–20) of histone H3 (B) compared with wild-type and mutant (H324L) human SUV39H1. Arrowheads mark the recombinant proteins. (C) Western analysis of nuclear extracts from wild-type and Su(var)3–9 null mutant [_Su(var)3–9_06/_Su(var)3–9_06] embryos with an α-dimethyl H3-K9 antibody. (D) Immunostaining of wild-type larval salivary gland chromosomes with α-dimethyl H3-K9 antibody. Whole chromosome set and enlargement of chromocentre region. Arrows denote the fourth chromosome. (E) Immuno staining of SU(VAR)3–9-deficient [_Su(var)3–9_06/_Su(var)3–9_06] larvae with α-dimethyl H3-K9 antibody. Arrows in enlarged chromocentre denotes the fourth chromosome. (F) Reconstitution of H3-K9 methylation in chromocentre heterochromatin by expression of SU(VAR)3–9–EGFP in Su(var)3–9 null mutant larvae. DNA staining with propidium iodide (D and E) and TOTO-3 (F).
Fig. 3. Yeast two-hybrid analysis of SU(VAR)3–9 interaction with HP1 and SU(VAR)3–7. Two-hybrid interaction tests of SU(VAR)3–9 baits with HP1 and SU(VAR)3–7 target constructs. LacZ activities are shown in parentheses. SU(VAR)3–9 regions are indicated, shaded for the overlapping 81 amino acid region with eIF2γ: yellow for the chromodomain, dark green for the SAC domain and green for the SET domain. In HP1 the chromodomain is indicated in yellow and the chromo-shadow domain in amber. Zinc fingers in SU(VAR)3–7 are indicated by white stripes.
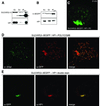
Fig. 4. In vivo and cytological analysis of interaction between SU(VAR)3–9 and HP1. (A and B) Immunoprecipitation of SU(VAR)3–9–EGFP (A) and HP1 (B) from heat-shocked transgenic and untreated wild-type 0–12 h embryos, respectively. Blots were probed with (A) α-SU(VAR)3–9, and (B) α-GFP (upper panels) and α-HP1 (lower panels). Ex, nuclear extract; Sn, supernatant; IP, immunoprecipitate. Arrowheads mark the signal of the antibody used for co-immunoprecipitation. (C) In vivo relocation of SU(VAR)3–9–EGFP by an HP1/PC chimeric protein. GFP fluorescence in a confocal section of a salivary gland nucleus. (D) Immunocytological analysis of transheterozygous larvae expressing SU(VAR)3–9–EGFP and the HP1/PC chimeric protein with α-GFP for SU(VAR)3–9–EGFP and α-β-gal for HP1/PC. (E) Double immunostaining for SU(VAR)3–9–EGFP with a mouse monoclonal α-GFP antibody and with a rabbit polyclonal α-HP1 antibody.
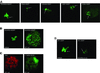
Fig. 5. Control of SU(VAR)3–9 and HP1 heterochromatin association. (A) Confocal sections of unfixed larval salivary gland nuclei expressing full-length SU(VAR)3–9–EGFP, SU(VAR)3–9NTerm–EGFP, SU(VAR)3–9Δchromo–EGFP, SU(VAR)3–9ΔSET–EGFP and SU(VAR)3–9TRX–EGFP protein variants in SU(VAR)3–9-deficient larvae. Arrows point to the fourth chromosome. (B) In vivo distribution of SU(VAR)3–9–EGFP in wild-type and HP1 deficient [Df(2L)TE128 × 11/_Su(var)2–5_04] salivary gland nuclei. (C) α-dimethyl H3-K9 antibody staining of an HP1-deficient salivary gland nucleus. DNA staining with propidium iodide. (D) In vivo analysis of HP1–EGFP heterochromatin association in wild-type and SU(VAR)3–9-deficient [_Su(var)3–9_06/_Su(var)3–9_06] nuclei of third instar larval salivary glands. Arrows point to the fourth chromosome.
Fig. 6. Genetic interactions between Su(var)3–9, Su(var)2–5 and Su(var)3–7 and the modifier effect of Y chromosome aneuploidy in _w_m4 PEV. (A) Heterozygotes for the loss-of-function mutant alleles _Su(var)3–9_06, _Su(var)2–5_05 and _Df(3R)AceD1_[Su(var)3–7 null] were combined with transgenic constructs introducing extra genomic copies of Su(var)3–9, Su(var)2–5 or Su(var)3–7, respectively. Resultant white variegation in offspring _w_m4 female is shown. (B) Genetic interaction to the PEV modifier effect of Y chromosome aneuploidy. First row shows white variegation in _w_m4/O males of +/+ (control) and of heterozygotes of the loss-of-function _Su(var)3–9_06, _Su(var)2–5_05 and _Df(3R)AceHD1_[Su(var)3–7 null] alleles, respectively. Second row shows white variegation in _w_m4/w/Y females of +/+ (control) and of heterozygotes with an additional genomic copy of Su(var)3–9, Su(var)2–5 and Su(var)3–7, respectively.
Fig. 7. Expression of human SUV39H1 in Drosophila. (A) Association of SUV39H1–EGFP with chromocentre heterochromatin and the fourth chromosome. In vivo confocal section of a salivary gland nucleus. (B) Relocation of SUV39H1–EGFP to euchromatic sites in transgenic lines with HP1/PC chimeric protein. (C and D) SUV39H1 rescues the dominant suppressor effect of Su(var)3–9 mutations: (C) control _w_m4/Y; _Su(var)3–9_06/+ and (D) _w_m4/Y; _Su(var)3–9_06/pP{GS[ry+, hs cDNA SUV39H1–EGFP]} males.
Similar articles
- SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing.
Schotta G, Ebert A, Reuter G. Schotta G, et al. Genetica. 2003 Mar;117(2-3):149-58. doi: 10.1023/a:1022923508198. Genetica. 2003. PMID: 12723694 Review. - Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila.
Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. Ebert A, et al. Genes Dev. 2004 Dec 1;18(23):2973-83. doi: 10.1101/gad.323004. Genes Dev. 2004. PMID: 15574598 Free PMC article. - Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location.
Greil F, van der Kraan I, Delrow J, Smothers JF, de Wit E, Bussemaker HJ, van Driel R, Henikoff S, van Steensel B. Greil F, et al. Genes Dev. 2003 Nov 15;17(22):2825-38. doi: 10.1101/gad.281503. Genes Dev. 2003. PMID: 14630943 Free PMC article. - Reduced levels of Su(var)3-9 but not Su(var)2-5 (HP1) counteract the effects on chromatin structure and viability in loss-of-function mutants of the JIL-1 histone H3S10 kinase.
Deng H, Bao X, Zhang W, Girton J, Johansen J, Johansen KM. Deng H, et al. Genetics. 2007 Sep;177(1):79-87. doi: 10.1534/genetics.107.075143. Epub 2007 Jul 29. Genetics. 2007. PMID: 17660558 Free PMC article. - Histone modification and the control of heterochromatic gene silencing in Drosophila.
Ebert A, Lein S, Schotta G, Reuter G. Ebert A, et al. Chromosome Res. 2006;14(4):377-92. doi: 10.1007/s10577-006-1066-1. Chromosome Res. 2006. PMID: 16821134 Review.
Cited by
- Epigenetic inheritance and gene expression regulation in early Drosophila embryos.
Ciabrelli F, Atinbayeva N, Pane A, Iovino N. Ciabrelli F, et al. EMBO Rep. 2024 Oct;25(10):4131-4152. doi: 10.1038/s44319-024-00245-z. Epub 2024 Sep 16. EMBO Rep. 2024. PMID: 39285248 Free PMC article. Review. - Prevalent Fast Evolution of Genes Involved in Heterochromatin Functions.
Lin L, Huang Y, McIntyre J, Chang CH, Colmenares S, Lee YCG. Lin L, et al. Mol Biol Evol. 2024 Sep 4;41(9):msae181. doi: 10.1093/molbev/msae181. Mol Biol Evol. 2024. PMID: 39189646 Free PMC article. - Role of diffusion and reaction of the constituents in spreading of histone modification marks.
Manivannan V, Inamdar MM, Padinhateeri R. Manivannan V, et al. PLoS Comput Biol. 2024 Jul 11;20(7):e1012235. doi: 10.1371/journal.pcbi.1012235. eCollection 2024 Jul. PLoS Comput Biol. 2024. PMID: 38991050 Free PMC article. - Genome-wide profiles of H3K9me3, H3K27me3 modifications, and DNA methylation during diapause of Asian corn borer (Ostrinia furnacalis).
Lv P, Yang X, Zhao X, Zhao Z, Du J. Lv P, et al. Genome Res. 2024 Jun 25;34(5):725-739. doi: 10.1101/gr.278661.123. Genome Res. 2024. PMID: 38866549 Free PMC article. - Inheritance of H3K9 methylation regulates genome architecture in Drosophila early embryos.
Atinbayeva N, Valent I, Zenk F, Loeser E, Rauer M, Herur S, Quarato P, Pyrowolakis G, Gomez-Auli A, Mittler G, Cecere G, Erhardt S, Tiana G, Zhan Y, Iovino N. Atinbayeva N, et al. EMBO J. 2024 Jul;43(13):2685-2714. doi: 10.1038/s44318-024-00127-z. Epub 2024 Jun 3. EMBO J. 2024. PMID: 38831123 Free PMC article.
References
- Alfageme C.R., Rudkin,G.T. and Cohen,L.H. (1980) Isolation, properties and cellular distribution of D1, a chromosomal protein of Drosophila. Chromosoma, 78, 1–31. - PubMed
- Bannister A.J., Zegermann,P., Patridge,J.F., Miska,E.A., Thomas,J.O., Allshire,T.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases