Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing - PubMed (original) (raw)
Comparative Study
Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing
Olga V Makarova et al. EMBO J. 2002.
Abstract
In each round of nuclear pre-mRNA splicing, the U4/U6*U5 tri-snRNP must be assembled from U4/U6 and U5 snRNPs, a reaction that is at present poorly understood. We have characterized a 61 kDa protein (61K) found in human U4/U6*U5 tri-snRNPs, which is homologous to yeast Prp31p, and show that it is required for this step. Immunodepletion of protein 61K from HeLa nuclear extracts inhibits tri-snRNP formation and subsequent spliceosome assembly and pre-mRNA splicing. Significantly, complementation with recombinant 61K protein restores each of these steps. Protein 61K is operationally defined as U4/U6 snRNP-specific as it remains bound to this particle at salt concentrations where the tri-snRNP dissociates. However, as shown by two-hybrid analysis and biochemical assays, protein 61K also interacts specifically with the U5 snRNP-associated 102K protein, indicating that it physically tethers U4/U6 to the U5 snRNP to yield the tri-snRNP. Interestingly, protein 61K is encoded by a gene (PRPF31) that has been shown to be linked to autosomal dominant retinitis pigmentosa. Thus, our studies suggest that disruptions in tri-snRNP formation and function resulting from mutations in the 61K protein may contribute to the manifestation of this disease.
Figures
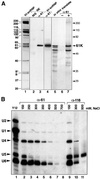
Fig. 1. The human 61K protein present in U4/U6·U5 tri-snRNPs is homologous to the yeast splicing factor Prp31p and the box C/D snoRNP proteins NOP56 and NOP58. (A) Alignment of the 61K protein sequence with S.cerevisiae Prp31p (DDBJ/EMBL/GenBank accession No. Z72876). (B) Alignment of the central part of the 61K protein encompassing amino acids 93–328 with the homologous sequences of the human NOP56 (aa 174–411, DDBJ/EMBL/GenBank accession No. Y12065) and NOP58 (aa 183–395, DDBJ/EMBL/GenBank accession No. AF123534) proteins. Sequence alignments were performed using the Clustal method.
Fig. 2. (A) Confirmation of the identity of the 61K cDNA. Lane 1: proteins of purified tri-snRNPs were separated by 10% SDS–PAGE and visualized by Coomassie Blue staining. The identity of the proteins is indicated on the left. Lanes 2–4: immunodetection of protein 61K in HeLa nuclear extract and tri-snRNPs. Proteins of purified U4/U6·U5 tri-snRNPs (lane 4) or HeLa nuclear extract (lanes 2 and 3) were separated by 10% SDS–PAGE, blotted onto a membrane and immunostained with affinity-purified anti-61K antibodies (lanes 3 and 4) or corresponding pre-immune serum (NIS, lane 2). Lanes 5–7: characterization of the protein generated by in vitro translation of the 61K protein cDNA. The 61K cDNA-derived translation product, labeled with [35S]methionine was immunoprecipitated with protein A– Sepharose-bound anti-61K antibodies; the bound material was fractionated by 10% SDS–PAGE and detected by fluorography (lane 7). Lanes 5 and 6 show an aliquot of the translation reaction and a mock immunoprecipitation performed in the absence of anti-61K antibodies, respectively. The position of molecular weight markers is indicated on the right. (B) Association of protein 61K with U4/U6 snRNPs. snRNP particles from HeLa nuclear extracts were immunoprecipitated with anti-61K (lanes 2–8) or anti-116K (lanes 9–11) antibodies, at the salt concentration indicated above each lane. Precipitation with the Sm-specific antibody Y12 at 150 mM NaCl (lane 1) served as a positive control. The co-precipitated RNAs were extracted, fractionated by denaturing 10% PAGE, transferred to a membrane and hybridized with probes specific for the U1, U2, U4, U5 and U6 snRNAs. The positions of the snRNAs are indicated on the left.
Fig. 3. Subcellular localization of the 61K protein in HeLa cells. HeLa cells were double-stained with affinity-purified anti-61K antibodies (A) and with monoclonal antibody Y12 (B). The red (A) and green fluorescence (B) was recorded independently and combined in an overlay image (C), leading to the yellow staining of structures decorated by both antibodies. The bars represent 10 µm.
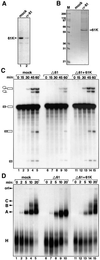
Fig. 4. Pre-mRNA splicing and spliceosome formation require the presence of the 61K protein. (A and B) Nuclear extract is efficiently immunodepleted of the 61K protein. (A) Western blot of mock-depleted (lane 1) or 61K-depleted extract (lane 2) probed with anti-61K antibody. (B) Material eluted from the respective columns with antigenic peptide was analyzed by 10% SDS–PAGE and stained with Coomassie Blue. (C) The time course of splicing of 32P-labeled pre-mRNA was monitored in mock-depleted (mock) and 61K-depleted (Δ61) extracts, or 61K-depleted extract complemented with recombinant 61K protein (Δ61+61K). Reaction products were analyzed on a denaturing polyacrylamide gel, and the splicing substrate/products detected by autoradiography. The positions of the bands corresponding to the pre-mRNA, intermediates and spliced products are indicated on the left. (D) Spliceosome assembly in mock-depleted extracts (mock), 61K-depleted extracts (Δ61) and 61K-depleted extracts complemented with the recombinant 61K (Δ61+61K) was analyzed by native gel electrophoresis followed by autoradiography. The bands corresponding to the H, A, B and C complexes, as well as the gel origin, are indicated on the left.
Fig. 5. Protein 61K is required for the formation of the U4/U6·U5 tri-snRNP. (A) Anti-Sm (α-Sm), anti-116K (α-116K) and anti-60K (α-60K) antibodies were used to immunoprecipitate snRNPs from mock-depleted (M), 61K-depleted (Δ) or 61K-depleted extract complemented with native protein 61K (Figure 4B) that was eluted from the anti-61K beads (Δ+). RNA was isolated from the immunoprecipitated snRNPs and characterized by northern blot analysis. (B and C) Destabilization of tri-snRNPs in the absence of protein 61K. Mock-depleted (B) or 61K-depleted (C) nuclear extract was fractionated on a 10–30% glycerol gradient (run in parallel). RNAs from each fraction were extracted, fractionated by denaturing 10% PAGE, and visualized by silver staining. The identities of the snRNAs (confirmed by northern hybridization, data not shown) are indicated on the right. For the mock-depleted extract, the distribution of protein 61K (detected by 10% SDS–PAGE, blotting onto a membrane and immunostaining with anti-61K) is shown in the lower part of (B).
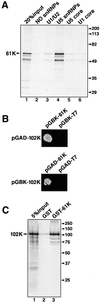
Fig. 6. (A) Protein 61K interacts with U5 snRNP. Protein 61K produced by translation in vitro was incubated with buffer only (lane 2), 12S U1 and U2 snRNPs (lane 3), U5 snRNPs (lane 4), or U5 and U1 core particles (lanes 5 and 6). The snRNP particles were then precipitated with the Sm-specific antibody Y12. The co-precipitated proteins were fractionated by SDS–PAGE and visualized by fluorography. (B) Protein 61K interacts with the U5-specific 102K protein in a yeast two-hybrid assay. The yeast strain AH109 was co-transformed with the indicated plasmid pairs and protein–protein interaction was identified on minimal medium lacking tryptophan, leucine, histidine and adenine. DNA-binding-domain vector (pGBKT7) and activation-domain vector (pGADT7) served as negative controls. (C) Co-precipitation of protein 102K with the 61K protein. 35S-labeled, in vitro translated protein 102K was incubated with GST (lane 2) or a protein 61K–GST fusion protein (lane 3). Proteins were precipitated using glutathione–Sepharose, fractionated by SDS–PAGE and visualized by fluorography.
Similar articles
- The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation.
Laggerbauer B, Liu S, Makarov E, Vornlocher HP, Makarova O, Ingelfinger D, Achsel T, Lührmann R. Laggerbauer B, et al. RNA. 2005 May;11(5):598-608. doi: 10.1261/rna.2300805. RNA. 2005. PMID: 15840814 Free PMC article. - The 20kD protein of human [U4/U6.U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins.
Teigelkamp S, Achsel T, Mundt C, Göthel SF, Cronshagen U, Lane WS, Marahiel M, Lührmann R. Teigelkamp S, et al. RNA. 1998 Feb;4(2):127-41. RNA. 1998. PMID: 9570313 Free PMC article. - Roles of the U5 snRNP in spliceosome dynamics and catalysis.
Turner IA, Norman CM, Churcher MJ, Newman AJ. Turner IA, et al. Biochem Soc Trans. 2004 Dec;32(Pt 6):928-31. doi: 10.1042/BST0320928. Biochem Soc Trans. 2004. PMID: 15506927 Review. - The assembly of a spliceosomal small nuclear ribonucleoprotein particle.
Patel SB, Bellini M. Patel SB, et al. Nucleic Acids Res. 2008 Nov;36(20):6482-93. doi: 10.1093/nar/gkn658. Epub 2008 Oct 14. Nucleic Acids Res. 2008. PMID: 18854356 Free PMC article. Review.
Cited by
- Three gene-targeted mouse models of RNA splicing factor RP show late-onset RPE and retinal degeneration.
Graziotto JJ, Farkas MH, Bujakowska K, Deramaudt BM, Zhang Q, Nandrot EF, Inglehearn CF, Bhattacharya SS, Pierce EA. Graziotto JJ, et al. Invest Ophthalmol Vis Sci. 2011 Jan 5;52(1):190-8. doi: 10.1167/iovs.10-5194. Print 2011 Jan. Invest Ophthalmol Vis Sci. 2011. PMID: 20811066 Free PMC article. - p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor.
Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A. Bell M, et al. EMBO J. 2002 Jun 3;21(11):2724-35. doi: 10.1093/emboj/21.11.2724. EMBO J. 2002. PMID: 12032085 Free PMC article. - Minor spliceosome components are predominantly localized in the nucleus.
Pessa HK, Will CL, Meng X, Schneider C, Watkins NJ, Perälä N, Nymark M, Turunen JJ, Lührmann R, Frilander MJ. Pessa HK, et al. Proc Natl Acad Sci U S A. 2008 Jun 24;105(25):8655-60. doi: 10.1073/pnas.0803646105. Epub 2008 Jun 16. Proc Natl Acad Sci U S A. 2008. PMID: 18559850 Free PMC article. - A guide to the biogenesis and functions of endogenous small non-coding RNAs in animals.
Jouravleva K, Zamore PD. Jouravleva K, et al. Nat Rev Mol Cell Biol. 2025 May;26(5):347-370. doi: 10.1038/s41580-024-00818-9. Epub 2025 Jan 24. Nat Rev Mol Cell Biol. 2025. PMID: 39856370 Review. - Variant snRNPs: New players within the spliceosome system.
Vazquez-Arango P, O'Reilly D. Vazquez-Arango P, et al. RNA Biol. 2018 Jan 2;15(1):17-25. doi: 10.1080/15476286.2017.1373238. Epub 2017 Oct 11. RNA Biol. 2018. PMID: 28876172 Free PMC article. Review.
References
- Behrens S.E. and Lührmann,R. (1991) Immunoaffinity purification of a [U4/U6·U5] tri-snRNP from human cells. Genes Dev., 5, 1439–1452. - PubMed
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases