Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans - PubMed (original) (raw)
Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans
Zezhang T Wen et al. Appl Environ Microbiol. 2002 Mar.
Erratum in
- Appl Environ Microbiol. 2003 Jan;69(1):722.
Abstract
Streptococcus mutans, the primary etiological agent of human dental caries, is an obligate biofilm-forming bacterium. The goals of this study were to identify the gene(s) required for biofilm formation by this organism and to elucidate the role(s) that some of the known global regulators of gene expression play in controlling biofilm formation. In S. mutans UA159, the brpA gene (for biofilm regulatory protein) was found to encode a novel protein of 406 amino acid residues. A strain carrying an insertionally inactivated copy of brpA formed longer chains than did the parental strain, aggregated in liquid culture, and was unable to form biofilms as shown by an in vitro biofilm assay. A putative homologue of the enzyme responsible for synthesis of autoinducer II (AI-2) of the bacterial quorum-sensing system was also identified in S. mutans UA159, but insertional inactivation of the gene (luxS(Sm)) did not alter colony or cell morphology or diminish the capacity of S. mutans to form biofilms. We also examined the role of the homologue of the Bacillus subtilis catabolite control protein CcpA in S. mutans in biofilm formation, and the results showed that loss of CcpA resulted in about a 60% decrease in the ability to form biofilms on an abiotic surface. From these data, we conclude that CcpA and BrpA may regulate genes that are required for stable biofilm formation by S. mutans.
Figures
FIG. 1.
Schematic diagram of the brpA locus and its flanking region. ORF1 and -2 encode hypothetical proteins; ORF4 codes for a protein of the histidine triad family; and ORF5 and ORF6 encode an ATP-binding protein and a permease of the ABC transporter system, respectively. Arrows represent the orientation of each individual ORF, and the numbers underneath indicate their respective positions in the chromosome.
FIG. 2.
Alignment of predicted amino acid sequence of BrpA of S. mutans (Sm) with the transcriptional regulator LytR of B. subtilis (Bs), and putative regulatory proteins CpsX of S. agalactiae (Sa), YeeG of L. lactis subsp. lactis (Lc), and Spy1733 of S. pyogenes (Sp). Identical residues are highlighted. Conserved regions of the protein are in boxes, and a consensus sequence is shown below these alignments. See the text for more detail.
FIG. 3.
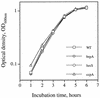
Growth curves of S. mutans UA159 (WT) and its derivatives. brpA, luxS, and ccpA refer to brpA, _luxS_Sm, and ccpA mutant strains, respectively. Cultures were grown in BM medium at 37°C in 5% CO2, and growth was monitored by measuring the absorbance of the cultures at 600 nm as described in Materials and Methods.
FIG. 4.
Morphological characteristics in BM medium. (A) Twenty-four-hour cultures of the _brpA_-deficient strain TW14 (tube 2) and its parent S. mutans UA159 (tube 1). (B and C) Phase-contrast microscopy of the 24-h cultures of UA159 and the brpA mutant, respectively (magnification, ×900). Data are representative of no fewer than three separate experiments.
FIG. 5.
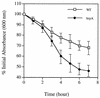
Autolysis of the whole cells of S. mutans UA159 (WT) and the _brpA_-deficient strain (brpA) grown in BHI medium. Mid-log-phase cultures (OD600 ≅ 0.7) were harvested by centrifugation, washed twice with ice-cold water, resuspended in 0.05 M Tris-HCl (pH 8.0) containing 0.2% Triton X-100, and incubated at 37°C with agitation (200 rpm). The changes of absorbance at 600 nm were determined as described in Materials and Methods. Data represent the average of three separate experiments, and the error bars indicate standard deviations.
FIG. 6.
Biofilm formation of S. mutans UA159 and its derivatives in BM medium. Crystal violet-stained 24-h biofilms of brpA (wells A5 to A7), ccpA (wells B4 and B5), and _luxS_Sm (well B3) mutants and their parental strain UA159 (wells A2 and A4 and well B1). Wells A1 and B2 are uninoculated BM medium as negative control. The graphs show quantitation of the biofilms formed after 6 h (left) and 24 h (right) by ccpA (ccpA), brpA (brpA), and _luxS_sm (luxS) mutants and the wild-type (WT) strains. See the text for more details. Data are representative of no fewer than three separate experiments. The error bars represent standard deviations.
Similar articles
- Streptococcus gordonii LuxS/autoinducer-2 quorum-sensing system modulates the dual-species biofilm formation with Streptococcus mutans.
Wang X, Li X, Ling J. Wang X, et al. J Basic Microbiol. 2017 Jul;57(7):605-616. doi: 10.1002/jobm.201700010. Epub 2017 May 9. J Basic Microbiol. 2017. PMID: 28485524 - Mutation of luxS affects biofilm formation in Streptococcus mutans.
Merritt J, Qi F, Goodman SD, Anderson MH, Shi W. Merritt J, et al. Infect Immun. 2003 Apr;71(4):1972-9. doi: 10.1128/IAI.71.4.1972-1979.2003. Infect Immun. 2003. PMID: 12654815 Free PMC article. - Deficiency of RgpG Causes Major Defects in Cell Division and Biofilm Formation, and Deficiency of LytR-CpsA-Psr Family Proteins Leads to Accumulation of Cell Wall Antigens in Culture Medium by Streptococcus mutans.
De A, Liao S, Bitoun JP, Roth R, Beatty WL, Wu H, Wen ZT. De A, et al. Appl Environ Microbiol. 2017 Aug 17;83(17):e00928-17. doi: 10.1128/AEM.00928-17. Print 2017 Sep 1. Appl Environ Microbiol. 2017. PMID: 28687645 Free PMC article. - Quorum sensing and biofilm formation by Streptococcus mutans.
Senadheera D, Cvitkovitch DG. Senadheera D, et al. Adv Exp Med Biol. 2008;631:178-88. doi: 10.1007/978-0-387-78885-2_12. Adv Exp Med Biol. 2008. PMID: 18792689 Review. - Polyketides/nonribosomal peptides from Streptococcus mutans and their ecological roles in dental biofilm.
Luo W, Zhang M, Zhou X, Xu X, Cheng X. Luo W, et al. Mol Oral Microbiol. 2024 Oct;39(5):261-269. doi: 10.1111/omi.12451. Epub 2024 Jan 11. Mol Oral Microbiol. 2024. PMID: 38212261 Review.
Cited by
- BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans.
Bitoun JP, Liao S, Yao X, Ahn SJ, Isoda R, Nguyen AH, Brady LJ, Burne RA, Abranches J, Wen ZT. Bitoun JP, et al. Appl Environ Microbiol. 2012 Apr;78(8):2914-22. doi: 10.1128/AEM.07823-11. Epub 2012 Feb 10. Appl Environ Microbiol. 2012. PMID: 22327589 Free PMC article. - Effects of oxygen on virulence traits of Streptococcus mutans.
Ahn SJ, Wen ZT, Burne RA. Ahn SJ, et al. J Bacteriol. 2007 Dec;189(23):8519-27. doi: 10.1128/JB.01180-07. Epub 2007 Oct 5. J Bacteriol. 2007. PMID: 17921307 Free PMC article. - A hypothetical protein of Streptococcus mutans is critical for biofilm formation.
Brown TA Jr, Ahn SJ, Frank RN, Chen YY, Lemos JA, Burne RA. Brown TA Jr, et al. Infect Immun. 2005 May;73(5):3147-51. doi: 10.1128/IAI.73.5.3147-3151.2005. Infect Immun. 2005. PMID: 15845523 Free PMC article. - Involvement of the adc operon and manganese homeostasis in Streptococcus gordonii biofilm formation.
Loo CY, Mitrakul K, Voss IB, Hughes CV, Ganeshkumar N. Loo CY, et al. J Bacteriol. 2003 May;185(9):2887-900. doi: 10.1128/JB.185.9.2887-2900.2003. J Bacteriol. 2003. PMID: 12700268 Free PMC article. - Deficiency of BrpB causes major defects in cell division, stress responses and biofilm formation by Streptococcus mutans.
Bitoun JP, Liao S, Xie GG, Beatty WL, Wen ZT. Bitoun JP, et al. Microbiology (Reading). 2014 Jan;160(Pt 1):67-78. doi: 10.1099/mic.0.072884-0. Epub 2013 Nov 4. Microbiology (Reading). 2014. PMID: 24190982 Free PMC article.
References
- Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signaling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. - PubMed
- Burne, R. A. 1998. Oral streptococci… products of their environment. J. Dent. Res. 77:445-452. - PubMed
- Burne, R. A. 1998. Regulation of gene expression in adherent population of oral streptococci, p. 55-71. In D. J. LeBlanc, M. S. Lantz, and L. M. Switalski (ed.), Microbial pathogenesis: current and emerging issues. Proceedings of the 2nd Annual Indiana Conference. Indiana University Press, Indianapolis, Ind.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources