VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum - PubMed (original) (raw)
Comparative Study
VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum
Antony Croxatto et al. J Bacteriol. 2002 Mar.
Abstract
Vibrio anguillarum possesses at least two N-acylhomoserine lactone (AHL) quorum-sensing circuits, one of which is related to the luxMN system of Vibrio harveyi. In this study, we have cloned an additional gene of this circuit, vanT, encoding a V. harveyi LuxR-like transcriptional regulator. A V. anguillarum Delta vanT null mutation resulted in a significant decrease in total protease activity due to loss of expression of the metalloprotease EmpA, but no changes in either AHL production or virulence. Additional genes positively regulated by VanT were identified from a plasmid-based gene library fused to a promoterless lacZ. Three lacZ fusions (serA::lacZ, hpdA-hgdA::lacZ, and sat-vps73::lacZ) were identified which exhibited decreased expression in the Delta vanT strain. SerA is similar to 3-phosphoglycerate dehydrogenases and catalyzes the first step in the serine-glycine biosynthesis pathway. HgdA has identity with homogentisate dioxygenases, and HpdA is homologous to 4-hydroxyphenylpyruvate dioxygenases (HPPDs) involved in pigment production. V. anguillarum strains require an active VanT to produce high levels of an L-tyrosine-induced brown color via HPPD, suggesting that VanT controls pigment production. Vps73 and Sat are related to Vibrio cholerae proteins encoded within a DNA locus required for biofilm formation. A V. anguillarum Delta vanT mutant and a mutant carrying a polar mutation in the sat-vps73 DNA locus were shown to produce defective biofilms. Hence, a new member of the V. harveyi LuxR transcriptional activator family has been characterized in V. anguillarum that positively regulates serine, metalloprotease, pigment, and biofilm production.
Figures
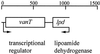
FIG. 1.
Genetic organization of the vanT DNA locus. Arrows indicate the direction of transcription.
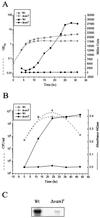
FIG. 2.
Analyses of metalloprotease activity in the wild type (Wt) and the vanT mutant. (A) Growth curve and expression of empA::lacZ transcriptional gene fusion. Overnight cultures were diluted to an OD600 nm of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (OD600, open symbols) and β-galactosidase expression (Miller units, solid symbols). Miller units for the vector control were between 100 and 200 U. (B) Protease activity. Overnight cultures were diluted to an OD600 of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (open symbols) and for azocasein degradation (OD442, solid symbols). For protease activity, 100 μl of filtered-sterilized culture supernatant was mixed with 100 μl of azocasein solution and incubated at 30°C for 2 h. The reaction was stopped by the addition of trichloroacetic acid, the unreacted azocasein was removed by centrifugation, and the absorbance at 442 nm was determined. (C) Northern analysis. Total RNA was isolated from 30-h cultures of the wild type and vanT mutant grown at 24°C and hybridized to a DNA fragment complementary to the empA gene.
FIG. 3.
Genetic organization of the cloned DNA fragments fused to the E. coli lacZ gene carried on pDM5-S5 and pDM5-S13. The horizontal arrows indicate the putative promoters and direction of transcription. The vertical arrow indicates the site of the plasmid insertion within the sat gene that created the polar mutation in the sat-vps73 DNA locus (strain DM70).
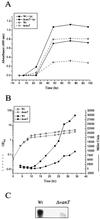
FIG. 4.
Measurement of pigment production and promoter activity of the hpdA gene. (A) For pigment production, overnight cultures of the wild type (Wt) and the vanT mutant containing pDM5-S13, which carries the hpdA gene and its promoter, were diluted to an OD600 of 0.05 into TSB medium with (solid symbols) and without 5 mM
l
-tyrosine (open symbols) and incubated with shaking at 24°C. Samples were taken from the culture at various times, and the supernatant was filter sterilized to remove bacteria. Pigment production in the supernatant was estimated by measuring the absorbance at 400 nm. (B) For hpdA promoter activity, an hpdA-lacZ transcriptional gene fusion was made and contained on pDM8-HpdA in the wild type and the vanT mutant. Overnight cultures were diluted into fresh TSB medium to an OD600 of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (OD600, open symbols) and β-galactosidase expression (Miller units, solid symbols). Miller units for the vector control were between 100 and 200 U. (C) Northern analysis. Total RNA was isolated from 30-h cultures of the wild type and vanT mutant grown at 24°C and hybridized to a DNA fragment complementary to the hpdA gene.
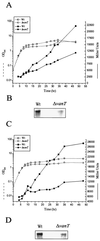
FIG. 5.
β-Galactosidase activity of vps73::lacZ and serA::lacZ transcriptional gene fusions. (A) vps73::lacZ was carried on pDM8-Vps73 and (C) serA::lacZ was carried on pDM8-SerA in the wild type (Wt) and vanT mutant. Overnight cultures were diluted into fresh TSB medium to an OD600 of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (OD600, open symbols) and β-galactosidase expression (Miller units, solid symbols). Miller units for the vector control were between 100 and 200 U. (B and D) Northern analysis. Total RNA was isolated from 30-h cultures of the wild type and vanT mutant grown at 24°C and hybridized to a DNA fragment complementary to either the vps73 gene (B) or the serA gene (D).
FIG. 6.
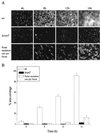
Biofilm attachment to a glass surface for the wild type (wt), the vanT mutant, and the mutant carrying a polar mutation in the sat-vps73 DNA locus. (A) Typical view of the progression of biofilm formation for each strain at 4 h, 8 h, 12 h, and 16 h. Slides were stained with acridine orange and then viewed with a fluorescent microscope. Bar, 20 μm. (B) Biofilm formation was quantified by determining the percent area coverage on a glass slide. For each strain, five images were taken at each time point and saved as a computer image. These images were converted into black (background) and white (biofilm) pixels using the Image Tool Software, version 2, and the percentage of white pixels is given as the percentage of biofilm covering the glass surface.
FIG. 7.
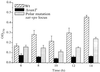
Quantification of bacteria in biofilms formed on PVC plastic by the wild-type (Wt) NB10, the vanT mutant, and the mutant carrying a polar mutation in the sat-vps73 DNA locus. Biofilms were allowed to form in a 96-well PVC microtiter dish. The microtiter dish was incubated for 6 h, 8 h, 10 h, 12 h, and 14 h at 24°C. The unattached bacteria were removed, and the biofilms were stained with crystal violet. The crystal violet was solubilized, and the absorbance, which correlated to the biofilm mass, was determined.
Similar articles
- A distinctive dual-channel quorum-sensing system operates in Vibrio anguillarum.
Croxatto A, Pride J, Hardman A, Williams P, Cámara M, Milton DL. Croxatto A, et al. Mol Microbiol. 2004 Jun;52(6):1677-89. doi: 10.1111/j.1365-2958.2004.04083.x. Mol Microbiol. 2004. PMID: 15186417 - RpoS induces expression of the Vibrio anguillarum quorum-sensing regulator VanT.
Weber B, Croxatto A, Chen C, Milton DL. Weber B, et al. Microbiology (Reading). 2008 Mar;154(Pt 3):767-780. doi: 10.1099/mic.0.2007/014167-0. Microbiology (Reading). 2008. PMID: 18310023 - The LuxM homologue VanM from Vibrio anguillarum directs the synthesis of N-(3-hydroxyhexanoyl)homoserine lactone and N-hexanoylhomoserine lactone.
Milton DL, Chalker VJ, Kirke D, Hardman A, Cámara M, Williams P. Milton DL, et al. J Bacteriol. 2001 Jun;183(12):3537-47. doi: 10.1128/JB.183.12.3537-3547.2001. J Bacteriol. 2001. PMID: 11371516 Free PMC article. - Gel shift analysis of the empA promoter region in Vibrio anguillarum.
Denkin SM, Sekaric P, Nelson DR. Denkin SM, et al. BMC Microbiol. 2004 Oct 29;4:42. doi: 10.1186/1471-2180-4-42. BMC Microbiol. 2004. PMID: 15516264 Free PMC article. - Quorum Sensing Gene Regulation by LuxR/HapR Master Regulators in Vibrios.
Ball AS, Chaparian RR, van Kessel JC. Ball AS, et al. J Bacteriol. 2017 Sep 5;199(19):e00105-17. doi: 10.1128/JB.00105-17. Print 2017 Oct 1. J Bacteriol. 2017. PMID: 28484045 Free PMC article. Review.
Cited by
- Bacterial quorum-sensing network architectures.
Ng WL, Bassler BL. Ng WL, et al. Annu Rev Genet. 2009;43:197-222. doi: 10.1146/annurev-genet-102108-134304. Annu Rev Genet. 2009. PMID: 19686078 Free PMC article. Review. - Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter clade.
Bruhn JB, Nielsen KF, Hjelm M, Hansen M, Bresciani J, Schulz S, Gram L. Bruhn JB, et al. Appl Environ Microbiol. 2005 Nov;71(11):7263-70. doi: 10.1128/AEM.71.11.7263-7270.2005. Appl Environ Microbiol. 2005. PMID: 16269767 Free PMC article. - Disruption of bacterial cell-to-cell communication by marine organisms and its relevance to aquaculture.
Natrah FM, Defoirdt T, Sorgeloos P, Bossier P. Natrah FM, et al. Mar Biotechnol (NY). 2011 Apr;13(2):109-26. doi: 10.1007/s10126-010-9346-3. Epub 2011 Jan 19. Mar Biotechnol (NY). 2011. PMID: 21246235 Review. - Evaluation of a new high-throughput method for identifying quorum quenching bacteria.
Tang K, Zhang Y, Yu M, Shi X, Coenye T, Bossier P, Zhang XH. Tang K, et al. Sci Rep. 2013 Oct 14;3:2935. doi: 10.1038/srep02935. Sci Rep. 2013. PMID: 24121744 Free PMC article. - The DmeRF System Is Involved in Maintaining Cobalt Homeostasis in Vibrio parahaemolyticus.
Zhao Y, Kong M, Yang J, Zhao X, Shi Y, Zhai Y, Qiu J, Zheng C. Zhao Y, et al. Int J Mol Sci. 2022 Dec 27;24(1):414. doi: 10.3390/ijms24010414. Int J Mol Sci. 2022. PMID: 36613858 Free PMC article.
References
- Actis, L. A., M. E. Tomalsky, and J. H. Crosa. 1999. Vibriosis, p. 523-557. In P. T. K. Woo and E. W. Bruno (ed.), Fish diseases and disorders, vol. 3: viral, bacterial and fungal infections. Cab International Publishing, Wallingford, United Kingdom.
- Agodi, A., S. Stefani, C. Corsaro, F. Campanile, S. Gribaldo, and G. Sichel. 1996. Study of a melanic pigment of Proteus mirabilis. Res. Microbiol. 147:167-174. - PubMed
- Austin, B., and D. A. Austin. 1999. Bacterial fish pathogens: disease of farmed and wild fish, p. 272-275. Springer and Praxis Publishing Ltd., Chichester, United Kingdom..
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials