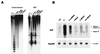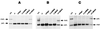Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality - PubMed (original) (raw)
Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality
Detlev Biniszkiewicz et al. Mol Cell Biol. 2002 Apr.
Abstract
Biallelic expression of Igf2 is frequently seen in cancers because Igf2 functions as a survival factor. In many tumors the activation of Igf2 expression has been correlated with de novo methylation of the imprinted region. We have compared the intrinsic susceptibilities of the imprinted region of Igf2 and H19, other imprinted genes, bulk genomic DNA, and repetitive retroviral sequences to Dnmt1 overexpression. At low Dnmt1 methyltransferase levels repetitive retroviral elements were methylated and silenced. The nonmethylated imprinted region of Igf2 and H19 was resistant to methylation at low Dnmt1 levels but became fully methylated when Dnmt1 was overexpressed from a bacterial artificial chromosome transgene. Methylation caused the activation of the silent Igf2 allele in wild-type and Dnmt1 knockout cells, leading to biallelic Igf2 expression. In contrast, the imprinted genes Igf2r, Peg3, Snrpn, and Grf1 were completely resistant to de novo methylation, even when Dnmt1 was overexpressed. Therefore, the intrinsic difference between the imprinted region of Igf2 and H19 and of other imprinted genes to postzygotic de novo methylation may be the molecular basis for the frequently observed de novo methylation and upregulation of Igf2 in neoplastic cells and tumors. Injection of Dnmt1-overexpressing embryonic stem cells in diploid or tetraploid blastocysts resulted in lethality of the embryo, which resembled embryonic lethality caused by Dnmt1 deficiency.
Figures
FIG. 1.
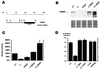
(A) Schematic diagram and restriction map of the BAC containing the Dnmt1 gene. Restriction sites are abbreviated as follows: N, _Not_I; P, _Pst_I. PCR screening of a mouse 129/Sv BAC library identified a BAC clone containing the Dnmt1 gene. The 150-kb BAC clone was shown to contain the complete Dnmt1 gene by a combination of PCR and Southern hybridization techniques (data not shown). (B) Western blot analysis of ES cell extracts using an antibody which was generated against the catalytic domain of Dnmt1. Similar results were obtained with a polyclonal antibody against the N-terminal region of Dnmt1 and are therefore not shown. All protein extracts were controlled by Coomassie-stained gel to ensure equal loading. (C) In vitro MTase activity assay measuring incorporation of the methyl group from [3H]_S_-adenosyl-
l
-methionine into a synthetic poly(dIdC) substrate. The in vitro assay was performed with three independently derived sets of protein extracts and their protein concentration was normalized. All results within a set were normalized to wild-type activity, which was defined as 100%. Each value represents the mean ± SEM. (D) 5-Methyl-cytosine content of genomic DNA from different ES cells and from differentiated tissues (liver, kidney), as measured by HPLC. Each value represents the mean ± SEM.
FIG. 2.
(A) Methylation-sensitive restriction enzyme assays with _Hpa_II using two repetitive probes, minor satellite centromeric repeat and an IAP. In these assays, low-molecular-weight bands indicate demethylated restriction sites, while high-molecular-weight bands indicate methylated restriction sites. (B) Northern blot analysis of IAP expression in differentiated cells. RNA loading was controlled for by hybridization with Gapdh.
FIG. 3.
The imprinted region of the genes Igf2r (A), Peg3 (B), and Grf1 (C) are resistant to de novo methylation by overexpression of Dnmt1 in ES cells. For the Igf2r gene, we assayed an MluI site in region 2, which resulted in a maternally methylated band and a paternally unmethylated band in wild-type ES cells (42). For the Peg3 gene, a _Sac_II site was assayed which is normally methylated on the maternal allele (25). A differentially methylated _Hha_I site was assayed for the Grf1 gene, which is paternally methylated in wild-type cells (34).
FIG. 4.
(A) Map of the upstream region of H19, which includes the DMD that regulates expression of Igf2 and H19. Location of the main imprinted region and the Southern probe (nt 1440 to 3332; GenBank accession no. U19619) are shown. Restriction sites are abbreviated as follows: E, _Eco_RI; S, _Sac_I. _Hha_I sites are indicated beneath the line. The region (nt 1089 to 1481; accession no. U19619) which was assayed by bisulfite modification is also indicated. (B) Methylation analysis of the DMD Igf2/H19 imprinted region, performed using the methylation-sensitive restriction enzyme _Hha_I and the methylation-insensitive enzyme _Sac_I. ES cells show one paternal methylated band and there are several undermethylated lower bands from the maternal allele. (C) Northern blot analysis of Igf2, H19, p57Kip2, and Igf2r expression in differentiated cells. RNA loading was controlled for by hybridization with Gapdh. The last two lanes on the right show a longer exposure of H19 expression only for the wild-type and Dnmt1+/+;BAC cells.
FIG.5.
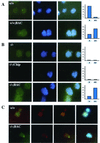
Igf2 RNA FISH in wild-type (A), Dnmt1+/+;BAC (A), Dnmt1−/− (B), Dnmt1chip/− (B), and Dnmt1−/−;BAC (B) cells. Content of images for each cell line is as follows, from left to right: Igf2 expression (green), nuclear 4′,6′-diamidino-2-phenylindole (DAPI) staining (blue), and merged image of Igf2 and DAPI. The graph to the right indicates the monoallelic versus biallelic Igf2 expression observed in each cell line. (C) Igf2 and H19 RNA FISH of wild-type and Dnmt1−/−;BAC cells. Images, from right to left, are Igf2 expression (green), H19 expression (red), a merged imaged of Igf2 and H19 expression, and a merged image of Igf2 and H19 expression with nuclear DAPI staining.
FIG. 6.
(A) Summary of the development (implantation) and survival of ES cell tetraploid blastocyst-derived mice or ES cell blastocyst-derived mice. Implantation is indicated in percent, while survial at dpc 14.5 is indicated in total numbers. ND, not determined. (B) Western blot analysis of mEF cell extracts using an antibody against the N-terminal domain of Dnmt1. All protein extracts were controlled by Coomassie-stained gel to ensure equal loading.
FIG. 7.
Summary of methylation and expression status of repetitive sequences such as IAP and of the imprinted genes Igf2r and Igf2 in cells with different levels of Dnmt1 expression, as measured by Western blot analysis (shown in second column). Three classes of genes (IAP, Igf2r, and Igf2) are subject to postzygotic de novo methylation at different levels of Dnmt1 expression, as illustrated. Repetitive IAP sequences are highly susceptible to de novo methylation. Igf2r and other imprinted genes, including Peg3, Snrpn, and Grf1 and the CpG islands of several nonimprinted genes, are completely resistant to de novo methylation. The imprinted region of Igf2 and H19 was resistant to de novo methylation at low Dnmt1 levels but became fully methylated at higher levels of Dnmt1 expression. The maternal and paternal alleles of Igf2 and Igf2r are indicated. The expression levels of IAP, Igf2r, and Igf2 are indicated by the number of + signs, while a − sign indicates no expression. Monoallelic or biallelic Igf2 expression in differentiated cells is indicated as the percentage of total _Igf2_-expressing cells. Embryonic survival of the ES cell tetraploid blastocyst-derived mice is summarized in the last column. Symbols: □, unmethylated; ▧, partially methylated (low); ▨, partially methylated (high); ▪, methylated; oval with one dot, monoallelic Igf2 expression; oval with two dots, biallelic Igf2 expression.
Similar articles
- Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes.
Tucker KL, Beard C, Dausmann J, Jackson-Grusby L, Laird PW, Lei H, Li E, Jaenisch R. Tucker KL, et al. Genes Dev. 1996 Apr 15;10(8):1008-20. doi: 10.1101/gad.10.8.1008. Genes Dev. 1996. PMID: 8608936 - Quantitative allele-specific expression and DNA methylation analysis of H19, IGF2 and IGF2R in the human placenta across gestation reveals H19 imprinting plasticity.
Buckberry S, Bianco-Miotto T, Hiendleder S, Roberts CT. Buckberry S, et al. PLoS One. 2012;7(12):e51210. doi: 10.1371/journal.pone.0051210. Epub 2012 Dec 5. PLoS One. 2012. PMID: 23227253 Free PMC article. - Domain-specific response of imprinted genes to reduced DNMT1.
Weaver JR, Sarkisian G, Krapp C, Mager J, Mann MR, Bartolomei MS. Weaver JR, et al. Mol Cell Biol. 2010 Aug;30(16):3916-28. doi: 10.1128/MCB.01278-09. Epub 2010 Jun 14. Mol Cell Biol. 2010. PMID: 20547750 Free PMC article. - Imprinting of the mouse Igf2r gene depends on an intronic CpG island.
Wutz A, Barlow DP. Wutz A, et al. Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
Cited by
- DNA hypomethylation induces a DNA replication-associated cell cycle arrest to block hepatic outgrowth in uhrf1 mutant zebrafish embryos.
Jacob V, Chernyavskaya Y, Chen X, Tan PS, Kent B, Hoshida Y, Sadler KC. Jacob V, et al. Development. 2015 Feb 1;142(3):510-21. doi: 10.1242/dev.115980. Epub 2015 Jan 6. Development. 2015. PMID: 25564650 Free PMC article. - Epigenetic regulation of fetal bone development and placental transfer of nutrients: progress for osteoporosis.
Bocheva G, Boyadjieva N. Bocheva G, et al. Interdiscip Toxicol. 2011 Dec;4(4):167-72. doi: 10.2478/v10102-011-0026-6. Interdiscip Toxicol. 2011. PMID: 22319250 Free PMC article. - Embryonic Stem Cells Lacking DNA Methyltransferases Differentiate into Neural Stem Cells that Are Defective in Self-Renewal.
Seo BJ, Hong TK, Yoon SH, Song JH, Uhm SJ, Song H, Hong K, Schöler HR, Do JT. Seo BJ, et al. Int J Stem Cells. 2023 Feb 28;16(1):44-51. doi: 10.15283/ijsc22138. Epub 2022 Oct 31. Int J Stem Cells. 2023. PMID: 36310027 Free PMC article. - UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma.
Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, Lachenmayer A, Revill K, Alsinet C, Sachidanandam R, Desai A, SenBanerjee S, Ukomadu C, Llovet JM, Sadler KC. Mudbhary R, et al. Cancer Cell. 2014 Feb 10;25(2):196-209. doi: 10.1016/j.ccr.2014.01.003. Epub 2014 Jan 30. Cancer Cell. 2014. PMID: 24486181 Free PMC article. - Alterations in gene expression and DNA methylation during murine and human lung alveolar septation.
Cuna A, Halloran B, Faye-Petersen O, Kelly D, Crossman DK, Cui X, Pandit K, Kaminski N, Bhattacharya S, Ahmad A, Mariani TJ, Ambalavanan N. Cuna A, et al. Am J Respir Cell Mol Biol. 2015 Jul;53(1):60-73. doi: 10.1165/rcmb.2014-0160OC. Am J Respir Cell Mol Biol. 2015. PMID: 25387348 Free PMC article.
References
- Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7:1663-1673. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous