GAL4 directs nucleosome sliding induced by NURF - PubMed (original) (raw)
GAL4 directs nucleosome sliding induced by NURF
Ju-Gyeong Kang et al. EMBO J. 2002.
Abstract
The Drosophila nucleosome remodeling factor (NURF) is an imitation switch (ISWI)-containing chromatin remodeling complex that can catalyze nucleosome repositioning at promoter regions to regulate access by the transcription machinery. Mononucleosomes reconstituted in vitro by salt dialysis adopt an ensemble of translational positions on DNA templates. NURF induces bi-directional 'sliding' of these nucleosomes to a subset of preferred positions. Here we show that mononucleosome sliding catalyzed by NURF bears similarity to nucleosome movement induced by elevated temperature. Moreover, we demonstrate that the GAL4 DNA-binding domain can extend NURF-induced nucleosome movement on a GAL4-E4 promoter, expanding the stretch of histone-free DNA at GAL4 recognition sites. The direction of NURF-induced nucleosome movement can be significantly modulated by asymmetric placement of tandem GAL4 sites relative to the nucleosome core particle. As such, sequence-specific, transcription factor-directed nucleosome sliding is likely to have substantial influence on promoter activation.
Figures
Fig. 1. Reconstituted nucleosomes occupy multiple positions on the GAL4-E4 promoter. (A) Native PAGE of mononucleosomes reconstituted on a 369 bp fragment carrying the GAL4-E4 promoter. Major nucleosome species are indicated as N1–N7′. Nucleosome positions and GAL4-binding sites determined by Exo III footprinting (precise to ± 2 bp) are illustrated on the right. The TATA box and restriction enzyme sites are indicated. (B) Exo III footprinting. Gel bands corresponding to individual N1–N7 nucleosome species (DNA was radiolabeled at either end of GAL4-E4 promoter) were excised, nucleosomes were eluted and digested with Exo III (400 U/ml) for 2 min at 37°C. DNAs were analyzed by electrophoresis in a 6% polyacrylamide gel containing 8 M urea. M indicates pBR322 _Hpa_II fragments as markers. Dots indicate major Exo III pauses. The Exo III footprint of GAL4 (–153 to –44) extends beyond the recognition sites (–140 to –56) (Carey et al., 1989; Carey and Smale, 2000).
Fig. 2. NURF mediates nucleosome sliding on the GAL4-E4 promoter. 369 bp GAL4-E4 mononucleosomes (40 nM) were incubated with NURF (0.4 nM) in the absence or presence of ATP for 30 min at 26°C (lanes 1 and 2), followed by native 4.5% PAGE in 0.5× TBE. N1, N2 and N4 nucleosomes were eluted from the gel slice and incubated with NURF and ATP as indicated, before electrophoresis on a second native gel (lanes 3–8).
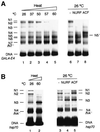
Fig. 3. Comparison of heat- and NURF-induced nucleosome sliding. Mononucleosomes reconstituted on GAL4-E4 (A) or hsp70 (B) radiolabeled DNAs were incubated for 30 min at the indicated temperatures in TE buffer and 1 mg/ml BSA. Samples were cooled on ice, and analyzed by native PAGE as in Figure 2 legend. Mononucleo somes were incubated with NURF (0.4 nM), [(A) lane 7; (B) lane 4] or ACF (1.6 nM), [(A) lane 8; (B) lane 5] in nucleosome sliding buffer at 26°C. Samples were electrophoresed as in Figure 2 legend.
Fig. 4. GAL4-DBD and NURF-induced extension of nucleosome sliding—Exo III footprinting analysis. Mononucleosomes were reconstituted on the end-labeled 369 bp GAL4-E4 promoter. Gel-purified nucleosomes (0.025 nM) and carrier DNA (20 µg/ml), N1 (lanes 1–6), N4 (lanes 7 and 8) were incubated with NURF (0.2 nM) and GAL4-DBD (0.3 nM) in the presence or absence of ATP as indicated. Reaction mixtures were digested by Exo III (400 U/ml) and analyzed on an 8 M urea–6% polyacrylamide gel. Dots indicate major Exo III pauses. Diagrams show the position of nucleosomes (open circles), GAL4 (filled ovals) and Exo III pauses (arrowheads).
Fig. 5. GAL4-DBD and NURF-induced extension of nucleosome sliding—MNase and restriction enzyme analysis. Nucleosomes were reconstituted on 32P uniformly labeled 369 bp GAL4-E4 fragment and incubated with GAL4 and/or NURF prior to MNase digestion. To map boundaries of MNase protection, DNA was purified and digested with _Ban_I (B) or _Fnu_4H1 (F). (A) Schematic illustration showing mononucleosomes and GAL4-DBD binding on 369 bp GAL4-E4 promoter. Arrowheads mark the boundaries of MNase protection. (B) Mononucleosome fractions (0.7 nM) purified by glycerol gradient centrifugation were incubated with NURF (0.2 nM) and/or GAL4 (30 nM monomer) for 30 min at 26°C, followed by MNase (25 U/ml) digestion (lanes 1–4). DNA was purified and further digested with _Ban_I (1300 U/ml, lanes 5 and 6) or _Fnu_4H1 (333 U/ml, lanes 7 and 8) and analyzed by 8% PAGE in TBE and autoradiography. Dots (filled and unfilled) indicate fragments protected by complexes of GAL4-DBD and N7 or N7′ nucleosomes, respectively. Binding of GAL4-DBD to free DNA or nucleosomes generates 109 and 145–240 bp MNase resistant fragments, respectively. In the absence of GAL4-DBD, MNase digestion of N1–N7 nucleosomes yields 145–155 bp fragments from core particles. (C) Mononucleosomes were incubated with GAL4(1–94) and GAL4(1–147) (20 nM monomer) in the presence or absence of NURF, and analyzed by MNase digestion and DNA gel electrophoresis. MNase cleavage at sites flanking the tandemly bound GAL4-DBDs appears to be more efficient for GAL4(1–94).
Fig. 6. Placement of GAL4 sites modulates direction of nucleosome sliding. (A) Gel-purified 369 bp GAL4-E4 mononucleosomes N2 (upper panel) and N4 (lower panel) were incubated with NURF in the presence of ATP and digested with MNase; DNA was purified, further digested with _Ban_I and _Fnu_4H1, and analyzed by PAGE and auto radiography as in the legend to Figure 5. The lower part of the gel (indicated by the box) was exposed to film ∼3 times longer than the upper part and contrast enhanced to visually reveal small DNA fragments. Quantitation of radioactive band intensity was performed by phosphoimager analysis. The percentage restriction enzyme cleavage was calculated by subtraction of the residual ∼150 bp nucleosome core particle DNA from an equivalent, uncleaved sample (lanes 1 and 4). Restriction fragments derived from N7 and N7′ nucleosomes are indicated by unfilled and filled dots, respectively. The 81 bp fragment derived from _Ban_I cleavage of N7′ core particle DNA is not visible at this exposure; note that the N7 and N7′ percentage values given in the text are approximate. (B) Gel-purified 369 bp GAL4-E4 mononucleo somes N2 (upper panel) and N4 (lower panel) were incubated with NURF and GAL4-DBD, in the presence or absence of ATP, and analyzed as above. Dots (unfilled and filled) indicate fragments corresponding to, or derived from, N7 and N7′ nucleosomes, respectively. The distribution of N7 and N7′ nucleosomes was calculated by phosphoimager analysis of radioactive fragments, and averaged from three sets of experiments.
Similar articles
- Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin.
Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Mizuguchi G, et al. Mol Cell. 1997 Dec;1(1):141-50. doi: 10.1016/s1097-2765(00)80015-5. Mol Cell. 1997. PMID: 9659911 - Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex.
Schwanbeck R, Xiao H, Wu C. Schwanbeck R, et al. J Biol Chem. 2004 Sep 17;279(38):39933-41. doi: 10.1074/jbc.M406060200. Epub 2004 Jul 15. J Biol Chem. 2004. PMID: 15262970 - Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions.
Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Xiao H, et al. Mol Cell. 2001 Sep;8(3):531-43. doi: 10.1016/s1097-2765(01)00345-8. Mol Cell. 2001. PMID: 11583616 - SWItched-on mobility.
Guschin D, Wolffe AP. Guschin D, et al. Curr Biol. 1999 Oct 7;9(19):R742-6. doi: 10.1016/s0960-9822(99)80473-4. Curr Biol. 1999. PMID: 10530996 Review. - Nucleosome remodeling factor NURF and in vitro transcription of chromatin.
Mizuguchi G, Wu C. Mizuguchi G, et al. Methods Mol Biol. 1999;119:333-42. doi: 10.1385/1-59259-681-9:333. Methods Mol Biol. 1999. PMID: 10804523 Review. No abstract available.
Cited by
- Asymmetry between the two acidic patches dictates the direction of nucleosome sliding by the ISWI chromatin remodeler.
Levendosky RF, Bowman GD. Levendosky RF, et al. Elife. 2019 May 16;8:e45472. doi: 10.7554/eLife.45472. Elife. 2019. PMID: 31094676 Free PMC article. - Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes.
Clapier CR, Iwasa J, Cairns BR, Peterson CL. Clapier CR, et al. Nat Rev Mol Cell Biol. 2017 Jul;18(7):407-422. doi: 10.1038/nrm.2017.26. Epub 2017 May 17. Nat Rev Mol Cell Biol. 2017. PMID: 28512350 Free PMC article. Review. - Interdomain Communication of the Chd1 Chromatin Remodeler across the DNA Gyres of the Nucleosome.
Nodelman IM, Bleichert F, Patel A, Ren R, Horvath KC, Berger JM, Bowman GD. Nodelman IM, et al. Mol Cell. 2017 Feb 2;65(3):447-459.e6. doi: 10.1016/j.molcel.2016.12.011. Epub 2017 Jan 19. Mol Cell. 2017. PMID: 28111016 Free PMC article. - The Chd1 chromatin remodeler shifts hexasomes unidirectionally.
Levendosky RF, Sabantsev A, Deindl S, Bowman GD. Levendosky RF, et al. Elife. 2016 Dec 29;5:e21356. doi: 10.7554/eLife.21356. Elife. 2016. PMID: 28032848 Free PMC article. - The Chd1 chromatin remodeler can sense both entry and exit sides of the nucleosome.
Nodelman IM, Horvath KC, Levendosky RF, Winger J, Ren R, Patel A, Li M, Wang MD, Roberts E, Bowman GD. Nodelman IM, et al. Nucleic Acids Res. 2016 Sep 19;44(16):7580-91. doi: 10.1093/nar/gkw406. Epub 2016 May 12. Nucleic Acids Res. 2016. PMID: 27174939 Free PMC article.
References
- Alfas J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials