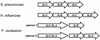Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586 - PubMed (original) (raw)
. 2002 Apr;184(7):2005-18.
doi: 10.1128/JB.184.7.2005-2018.2002.
Iain Anderson, Natalia Ivanova, Gary Reznik, Tamara Los, Athanasios Lykidis, Anamitra Bhattacharyya, Allen Bartman, Warren Gardner, Galina Grechkin, Lihua Zhu, Olga Vasieva, Lien Chu, Yakov Kogan, Oleg Chaga, Eugene Goltsman, Axel Bernal, Niels Larsen, Mark D'Souza, Theresa Walunas, Gordon Pusch, Robert Haselkorn, Michael Fonstein, Nikos Kyrpides, Ross Overbeek
Affiliations
- PMID: 11889109
- PMCID: PMC134920
- DOI: 10.1128/JB.184.7.2005-2018.2002
Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586
Vinayak Kapatral et al. J Bacteriol. 2002 Apr.
Abstract
We present a complete DNA sequence and metabolic analysis of the dominant oral bacterium Fusobacterium nucleatum. Although not considered a major dental pathogen on its own, this anaerobe facilitates the aggregation and establishment of several other species including the dental pathogens Porphyromonas gingivalis and Bacteroides forsythus. The F. nucleatum strain ATCC 25586 genome was assembled from shotgun sequences and analyzed using the ERGO bioinformatics suite (http://www.integratedgenomics.com). The genome contains 2.17 Mb encoding 2,067 open reading frames, organized on a single circular chromosome with 27% GC content. Despite its taxonomic position among the gram-negative bacteria, several features of its core metabolism are similar to that of gram-positive Clostridium spp., Enterococcus spp., and Lactococcus spp. The genome analysis has revealed several key aspects of the pathways of organic acid, amino acid, carbohydrate, and lipid metabolism. Nine very-high-molecular-weight outer membrane proteins are predicted from the sequence, none of which has been reported in the literature. More than 137 transporters for the uptake of a variety of substrates such as peptides, sugars, metal ions, and cofactors have been identified. Biosynthetic pathways exist for only three amino acids: glutamate, aspartate, and asparagine. The remaining amino acids are imported as such or as di- or oligopeptides that are subsequently degraded in the cytoplasm. A principal source of energy appears to be the fermentation of glutamate to butyrate. Additionally, desulfuration of cysteine and methionine yields ammonia, H(2)S, methyl mercaptan, and butyrate, which are capable of arresting fibroblast growth, thus preventing wound healing and aiding penetration of the gingival epithelium. The metabolic capabilities of F. nucleatum revealed by its genome are therefore consistent with its specialized niche in the mouth.
Figures
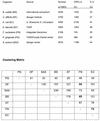
FIG. 1.
Comparisons of chromosomal clustering (CC) of ORFs in F. nucleatum strain ATCC 25586 with B. subtilis, C. difficile, E. coli, E. faecalis, P. gingivalis, and S. aureus EMRSA. Clustering is calculated by searching for pairs of close bidirectional best hits on a run of ORFs on the same strand using the ERGO chromosomal clustering tool. The number of clusters and the comparative matrix are represented below.
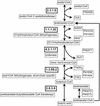
FIG. 2.
Metabolic reconstruction of the butyric acid fermentation pathway in F. nucleatum. All the ORFs that code for enzymes involved in the pathway have been identified.
FIG. 3.
An overview of amino acid synthesis (A) and degradation (B) in F. nucleatum. Orange, brick red, or gold boxes indicate that the pathway is present. A gray box indicates a missing pathway, and the striped box indicates that a few candidates of the pathway are present.
FIG. 3.
An overview of amino acid synthesis (A) and degradation (B) in F. nucleatum. Orange, brick red, or gold boxes indicate that the pathway is present. A gray box indicates a missing pathway, and the striped box indicates that a few candidates of the pathway are present.
FIG. 4.
Comparison of the lic1 operon of F. nucleatum with that of S. pneumoniae and H. influenzae. F. nucleatum has two operons that resemble the lic1 loci in the other bacteria. Operon 1 consists of two ORFs (FN0110 and FN0111), while operon 2 consists of three ORFs (FN1668, FN1669, and FN1670). The F. nucleatum choline kinase and phosphocholine cytidylyl-transferase genes are fused and presumably form two bifunctional enzymes. One of these operons might have arisen by gene duplication in F. nucleatum more recently than the gene fusion event.
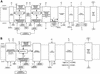
FIG. 5.
(A) Representation of the aerobic respiration pathway and proton flow in F. nucleatum. (B) Representation of the anaerobic respiration pathway in F. nucleatum.
FIG. 6.
(Upper panel) Clustering of hemolysin genes (red arrow) of F. nucleatum with those of N. meningitidis, E. coli O157:H7, and P. multocida. Except in the case of E. coli O157:H7, the ORF for hemolysin activator (green arrow) is upstream of the hemolysin gene. The broken lines indicate the presence of insertion elements. The“pinned region” algorithm was used to describe the pattern organization. The yellow, brown, and gray arrows represent ORFs and operons that are not conserved in this hemolysin cluster. (Lower panel) Clustering of a second hemolysin gene (red arrow) of F. nucleatum with those of B. cepacia and Methylobacillus flagellatus. This hemolysin gene in F. nucleatum is truncated.
Similar articles
- Genome analysis of F. nucleatum sub spp vincentii and its comparison with the genome of F. nucleatum ATCC 25586.
Kapatral V, Ivanova N, Anderson I, Reznik G, Bhattacharyya A, Gardner WL, Mikhailova N, Lapidus A, Larsen N, D'Souza M, Walunas T, Haselkorn R, Overbeek R, Kyrpides N. Kapatral V, et al. Genome Res. 2003 Jun;13(6A):1180-9. doi: 10.1101/gr.566003. Genome Res. 2003. PMID: 12799352 Free PMC article. - Genome sequence of Fusobacterium nucleatum subspecies polymorphum - a genetically tractable fusobacterium.
Karpathy SE, Qin X, Gioia J, Jiang H, Liu Y, Petrosino JF, Yerrapragada S, Fox GE, Haake SK, Weinstock GM, Highlander SK. Karpathy SE, et al. PLoS One. 2007 Aug 1;2(7):e659. doi: 10.1371/journal.pone.0000659. PLoS One. 2007. PMID: 17668047 Free PMC article. - Fusobacterium Genomics Using MinION and Illumina Sequencing Enables Genome Completion and Correction.
Todd SM, Settlage RE, Lahmers KK, Slade DJ. Todd SM, et al. mSphere. 2018 Jul 5;3(4):e00269-18. doi: 10.1128/mSphere.00269-18. mSphere. 2018. PMID: 29976645 Free PMC article. - Halitosis vaccines targeting FomA, a biofilm-bridging protein of fusobacteria nucleatum.
Liu PF, Huang IF, Shu CW, Huang CM. Liu PF, et al. Curr Mol Med. 2013 Sep;13(8):1358-67. doi: 10.2174/15665240113139990063. Curr Mol Med. 2013. PMID: 23865430 Review. - Genomics of oral bacteria.
Duncan MJ. Duncan MJ. Crit Rev Oral Biol Med. 2003;14(3):175-87. doi: 10.1177/154411130301400303. Crit Rev Oral Biol Med. 2003. PMID: 12799321 Review.
Cited by
- Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa.
Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, Hackstadt T. Ellison DW, et al. Infect Immun. 2008 Feb;76(2):542-50. doi: 10.1128/IAI.00952-07. Epub 2007 Nov 19. Infect Immun. 2008. PMID: 18025092 Free PMC article. - Characterization of aid1, a novel gene involved in Fusobacterium nucleatum interspecies interactions.
Kaplan A, Kaplan CW, He X, McHardy I, Shi W, Lux R. Kaplan A, et al. Microb Ecol. 2014 Aug;68(2):379-87. doi: 10.1007/s00248-014-0400-y. Epub 2014 Mar 20. Microb Ecol. 2014. PMID: 24643713 Free PMC article. - Characterization of the non-glandular gastric region microbiota in Helicobacter suis-infected versus non-infected pigs identifies a potential role for Fusobacterium gastrosuis in gastric ulceration.
De Witte C, Demeyere K, De Bruyckere S, Taminiau B, Daube G, Ducatelle R, Meyer E, Haesebrouck F. De Witte C, et al. Vet Res. 2019 May 24;50(1):39. doi: 10.1186/s13567-019-0656-9. Vet Res. 2019. PMID: 31126330 Free PMC article. - Is a fusobacterium nucleatum infection in the colon a risk factor for colorectal cancer?: a systematic review and meta-analysis protocol.
Idrissi Janati A, Karp I, Sabri H, Emami E. Idrissi Janati A, et al. Syst Rev. 2019 May 10;8(1):114. doi: 10.1186/s13643-019-1031-7. Syst Rev. 2019. PMID: 31077259 Free PMC article. - Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch.
Serganov A, Huang L, Patel DJ. Serganov A, et al. Nature. 2009 Mar 12;458(7235):233-7. doi: 10.1038/nature07642. Epub 2009 Jan 25. Nature. 2009. PMID: 19169240 Free PMC article.
References
- Andersen, R. N., N. Ganeshkumar, and P. E. Kolenbrander. 1998. Helicobacter pylori adheres selectively to Fusobacterium spp. Oral Microbiol. Immunol. 13:51-54. - PubMed
- Beatrix, B., K. Bendrat, S. Rospert, and W. Buckel. 1990. The biotin-dependent sodium ion pump glutaconyl-CoA decarboxylase from Fusobacterium nucleatum (subsp. nucleatum). Comparison with the glutaconyl-CoA decarboxylases from Gram-positive bacteria. Arch. Microbiol. 154:362-369. - PubMed
- Bolstad, A. I. 1994. Sizing of the Fusobacterium nucleatum genome by pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 123:145-152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous