Structural studies of the scrapie prion protein by electron crystallography - PubMed (original) (raw)
Structural studies of the scrapie prion protein by electron crystallography
Holger Wille et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Because the insolubility of the scrapie prion protein (PrP(Sc)) has frustrated structural studies by x-ray crystallography or NMR spectroscopy, we used electron crystallography to characterize the structure of two infectious variants of the prion protein. Isomorphous two-dimensional crystals of the N-terminally truncated PrP(Sc) (PrP 27-30) and a miniprion (PrP(Sc)106) were identified by negative stain electron microscopy. Image processing allowed the extraction of limited structural information to 7 A resolution. By comparing projection maps of PrP 27-30 and PrP(Sc)106, we visualized the 36-residue internal deletion of the miniprion and localized the N-linked sugars. The dimensions of the monomer and the locations of the deleted segment and sugars were used as constraints in the construction of models for PrP(Sc). Only models featuring parallel beta-helices as the key element could satisfy the constraints. These low-resolution projection maps and models have implications for understanding prion propagation and the pathogenesis of neurodegeneration.
Figures
Figure 1
2D crystals of PrP 27-30. (A) A 2D crystal of PrP 27-30 stained with 2% uranyl acetate showing an apparent hexagonal lattice. (B) High power view of a crystal after CTF correction and several rounds of correlation-mapping and averaging. (C) Section of a power spectrum after averaging showing spots out to the 11th order, corresponding to ≈7 Å (arrow). (D) Crystallographic averaging further improved the amount of detail visible. A p3 plane group was used. (E) Typical prion rod with an aggregate of “crystal” subunits at each end. Some protofilaments reveal rows of dense stain accumulations, suggesting stacked subunits (arrowheads). [Bars = 100 nm.]
Figure 2
Nanogold labeling of the N-linked sugars. (A) Uranyl acetate-stained 2D crystal of Nanogold-labeled PrP 27-30. The high contrast of the uranyl stain obscures some of the labels, but others are clearly visible (arrowheads). [Bar = 100 nm.] (B) Image processing result of a labeled crystal after correlation-mapping and averaging followed by crystallographic averaging. (C) Subtraction map between labeled and unlabeled crystals showing major differences in lighter shades. (D) Overlay of the statistically significant differences calculated from C in yellow onto a projection map of PrP 27-30.
Figure 3
2D crystals of PrPSc106. (A) A 2D crystal of PrPSc106 stained with uranyl acetate. [Bar = 100 nm.] (B) Image processing result after correlation-mapping and averaging followed by crystallographic averaging. (C and D) Subtraction maps between the averages of PrP 27-30 (Fig. 1_D_) and PrPSc106 (B). (C) PrPSc106 minus PrP 27-30 and (D) PrP 27-30 minus PrPSc106, showing major differences in lighter shades. (E and F) The statistically significant differences between PrP 27-30 and PrPSc106 calculated from C and D in red and blue, respectively, overlaid onto the crystallographic average of PrP 27-30 (Fig. 1_D_).
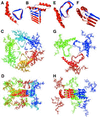
Figure 4
β-Helical models of PrP 27-30. (A and B) Top and side views, respectively, of PrP 27-30 modeled with a left-handed β-helix. The β-helical portion of the model is based on the Methanosarcina thermophila γ-carbonic anhydrase structure. (C and D) Top and side views, respectively, of the trimer of dimer model of PrP 27-30 with left-handed β-helices. (E and F) Top and side views, respectively, of PrP 27-30 modeled with a right-handed β-helix. The β-helical portion of the model is based on the most regular helical turns of Bordetella pertussis P.69 pertactin. (G and H) Top and side views, respectively, of the trimer model of PrP 27-30 with right-handed β-helices. The structure of the α-helices was derived from the solution structure of recombinant hamster PrP (–13). In the single-molecule images (A, B, E, and F), residues 141–176 that are deleted in PrP106 are colored blue.
Similar articles
- Electron crystallography of the scrapie prion protein complexed with heavy metals.
Wille H, Govaerts C, Borovinskiy A, Latawiec D, Downing KH, Cohen FE, Prusiner SB. Wille H, et al. Arch Biochem Biophys. 2007 Nov 15;467(2):239-48. doi: 10.1016/j.abb.2007.08.010. Epub 2007 Aug 23. Arch Biochem Biophys. 2007. PMID: 17935686 Free PMC article. - Ultrastructural studies on scrapie prion protein crystals obtained from reverse micellar solutions.
Wille H, Prusiner SB. Wille H, et al. Biophys J. 1999 Feb;76(2):1048-62. doi: 10.1016/S0006-3495(99)77270-X. Biophys J. 1999. PMID: 9916037 Free PMC article. - Biochemistry and structure of PrP(C) and PrP(Sc).
Riesner D. Riesner D. Br Med Bull. 2003;66:21-33. doi: 10.1093/bmb/66.1.21. Br Med Bull. 2003. PMID: 14522846 Review. - X-ray diffraction of scrapie prion rods and PrP peptides.
Nguyen JT, Inouye H, Baldwin MA, Fletterick RJ, Cohen FE, Prusiner SB, Kirschner DA. Nguyen JT, et al. J Mol Biol. 1995 Sep 29;252(4):412-22. doi: 10.1006/jmbi.1995.0507. J Mol Biol. 1995. PMID: 7563061 - Transition of the prion protein from a structured cellular form (PrPC ) to the infectious scrapie agent (PrPSc ).
Baral PK, Yin J, Aguzzi A, James MNG. Baral PK, et al. Protein Sci. 2019 Dec;28(12):2055-2063. doi: 10.1002/pro.3735. Epub 2019 Oct 25. Protein Sci. 2019. PMID: 31583788 Free PMC article. Review.
Cited by
- Structural insight into conformational change in prion protein by breakage of electrostatic network around H187 due to its protonation.
Lee J, Chang I. Lee J, et al. Sci Rep. 2019 Dec 17;9(1):19305. doi: 10.1038/s41598-019-55808-1. Sci Rep. 2019. PMID: 31848406 Free PMC article. - Mutant PrPSc conformers induced by a synthetic peptide and several prion strains.
Tremblay P, Ball HL, Kaneko K, Groth D, Hegde RS, Cohen FE, DeArmond SJ, Prusiner SB, Safar JG. Tremblay P, et al. J Virol. 2004 Feb;78(4):2088-99. doi: 10.1128/jvi.78.4.2088-2099.2004. J Virol. 2004. PMID: 14747574 Free PMC article. - From conversion to aggregation: protofibril formation of the prion protein.
DeMarco ML, Daggett V. DeMarco ML, et al. Proc Natl Acad Sci U S A. 2004 Feb 24;101(8):2293-8. doi: 10.1073/pnas.0307178101. Proc Natl Acad Sci U S A. 2004. PMID: 14983003 Free PMC article. - Conformational polymorphism of the amyloidogenic peptide homologous to residues 113-127 of the prion protein.
Satheeshkumar KS, Jayakumar R. Satheeshkumar KS, et al. Biophys J. 2003 Jul;85(1):473-83. doi: 10.1016/S0006-3495(03)74492-0. Biophys J. 2003. PMID: 12829502 Free PMC article. - Misfolding pathways of the prion protein probed by molecular dynamics simulations.
Barducci A, Chelli R, Procacci P, Schettino V. Barducci A, et al. Biophys J. 2005 Feb;88(2):1334-43. doi: 10.1529/biophysj.104.049882. Epub 2004 Nov 19. Biophys J. 2005. PMID: 15556981 Free PMC article.
References
- Cohen F E, Prusiner S B. In: Prion Biology and Diseases. Prusiner S B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 191–228.
- Prusiner S B, McKinley M P, Bowman K A, Bolton D C, Bendheim P E, Groth D F, Glenner G G. Cell. 1983;35:349–358. - PubMed
- Nguyen J T, Inouye H, Baldwin M A, Fletterick R J, Cohen F E, Prusiner S B, Kirschner D A. J Mol Biol. 1995;252:412–422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials