Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster - PubMed (original) (raw)
Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster
Wen Wang et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Non-protein-coding RNA genes play an important role in various biological processes. How new RNA genes originated and whether this process is controlled by similar evolutionary mechanisms for the origin of protein-coding genes remains unclear. A young chimeric RNA gene that we term sphinx (spx) provides the first insight into the early stage of evolution of RNA genes. spx originated as an insertion of a retroposed sequence of the ATP synthase chain F gene at the cytological region 60DB since the divergence of Drosophila melanogaster from its sibling species 2-3 million years ago. This retrosequence, which is located at 102F on the fourth chromosome, recruited a nearby exon and intron, thereby evolving a chimeric gene structure. This molecular process suggests that the mechanism of exon shuffling, which can generate protein-coding genes, also plays a role in the origin of RNA genes. The subsequent evolutionary process of spx has been associated with a high nucleotide substitution rate, possibly driven by a continuous positive Darwinian selection for a novel function, as is shown in its sex- and development-specific alternative splicing. To test whether spx has adapted to different environments, we investigated its population genetic structure in the unique "Evolution Canyon" in Israel, revealing a similar haplotype structure in spx, and thus similar evolutionary forces operating on spx between environments.
Figures
Figure 1
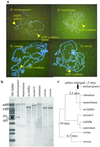
(a) FISH results showing that there are two red signals (arrows) in D. melanogaster. (b) Southern hybridization results also show that D. melanogaster has two copies homologous to the probe, which is the cDNA of ATP synthase chain F gene, but the other species have only one. (c) Phylogenetic tree of D. melanogaster subgroup (36). Divergence time of some nodes and emergence time of sphinx is indicated.
Figure 2
Alignment of the sphinx locus sequence of D. melanogaster with the sequences of the correspondent region of D. sechellia, CG4692 cDNA of D. melanogaster, and deduced partial cDNA of D. simulans. Asterisk indicates the start base of spx transcripts, dashes indicate deletions, and dots show the identical bases. The two exon sequences of the male-specific transcript (_sphinx_-m) are in capital letters. The two alternative adenylation signals (AATAAA) are in bold. All of the splicing sites (gt or cag) are indicated by boldface and vertical arrows. The retroposed sequence is flanked by the short direct repeats of TTCG, which are double underlined and indicated by the horizontal arrows. The region homologous to the ATP synthase gene is underlined. The remaining retroposed sequence is homologous to the terminal inverted repeat of S element, and therefore, the recipient slicing site was provided by the S element sequence.
Figure 3
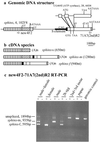
(a) Gene structure of sphinx and its parental gene, ATP synthase chain F. Blank blocks represent the exon of the ATP synthase gene. Black blocks represent the terminal inverted repeat sequence of S element, a small part (27 bp) of which is recruited into the second exon of sphinx. Other stripped blocks are exon sequences that are endogenous in the fourth chromosome. Some important changes in the retroposed ATP synthase sequence are indicated, including the change of start codon into GTG, introduction of a stop codon, and three deletions (d1 = 42 bp, d2 = 1 bp, d3 = 4 bp). The positions of the two primers used for RT-PCR are indicated by arrows. Splicing sites are marked by GT or CAG. Polyadenylation sites are marked by AATAAA. (b) mRNA species resulted from alternative adenylation and splicing. Sphinx-m is detected in males and eggs, sphinx-f is in females and eggs, _sphinx_-s is in both females and males, and the unspliced one is in males, larvae, and pupae. (c) RT-PCR results. “−” indicates the RT-PCR-negative controls in which everything is the same as the positive (+) except omitting reverse transcriptase. The last lane is a positive control using genomic DNA as the template. The primer locations on the gene are shown in a.
Similar articles
- Origin of new genes and source for N-terminal domain of the chimerical gene, jingwei, in Drosophila.
Long M, Wang W, Zhang J. Long M, et al. Gene. 1999 Sep 30;238(1):135-41. doi: 10.1016/s0378-1119(99)00229-2. Gene. 1999. PMID: 10570991 - Origin of new genes: evidence from experimental and computational analyses.
Long M, Deutsch M, Wang W, Betrán E, Brunet FG, Zhang J. Long M, et al. Genetica. 2003 Jul;118(2-3):171-82. Genetica. 2003. PMID: 12868607 Review. - Evolution of hydra, a recently evolved testis-expressed gene with nine alternative first exons in Drosophila melanogaster.
Chen ST, Cheng HC, Barbash DA, Yang HP. Chen ST, et al. PLoS Genet. 2007 Jul;3(7):e107. doi: 10.1371/journal.pgen.0030107. PLoS Genet. 2007. PMID: 17616977 Free PMC article. - [Evolutionary fate and expression patterns of chimeric new genes in Drosophila melanogaster].
Zhan ZB, Zhang Y, Zhao RP, Wang W. Zhan ZB, et al. Dongwuxue Yanjiu. 2011 Dec;32(6):585-95. doi: 10.3724/SP.J.1141.2011.06585. Dongwuxue Yanjiu. 2011. PMID: 22184016 Chinese. - Duplication-degeneration as a mechanism of gene fission and the origin of new genes in Drosophila species.
Wang W, Yu H, Long M. Wang W, et al. Nat Genet. 2004 May;36(5):523-7. doi: 10.1038/ng1338. Epub 2004 Apr 4. Nat Genet. 2004. PMID: 15064762
Cited by
- Adaptive evolution and the birth of CTCF binding sites in the Drosophila genome.
Ni X, Zhang YE, Nègre N, Chen S, Long M, White KP. Ni X, et al. PLoS Biol. 2012;10(11):e1001420. doi: 10.1371/journal.pbio.1001420. Epub 2012 Nov 6. PLoS Biol. 2012. PMID: 23139640 Free PMC article. - Two primate-specific small non-protein-coding RNAs in transgenic mice: neuronal expression, subcellular localization and binding partners.
Khanam T, Rozhdestvensky TS, Bundman M, Galiveti CR, Handel S, Sukonina V, Jordan U, Brosius J, Skryabin BV. Khanam T, et al. Nucleic Acids Res. 2007;35(2):529-39. doi: 10.1093/nar/gkl1082. Epub 2006 Dec 14. Nucleic Acids Res. 2007. PMID: 17175535 Free PMC article. - A new retroposed gene in Drosophila heterochromatin detected by microarray-based comparative genomic hybridization.
Fan C, Long M. Fan C, et al. J Mol Evol. 2007 Feb;64(2):272-83. doi: 10.1007/s00239-006-0169-9. Epub 2006 Dec 19. J Mol Evol. 2007. PMID: 17177089 - Gene duplication and other evolutionary strategies: from the RNA world to the future.
Brosius J. Brosius J. J Struct Funct Genomics. 2003;3(1-4):1-17. doi: 10.1023/a:1022627311114. J Struct Funct Genomics. 2003. PMID: 12836680 Review. - Extensive structural renovation of retrogenes in the evolution of the Populus genome.
Zhu Z, Zhang Y, Long M. Zhu Z, et al. Plant Physiol. 2009 Dec;151(4):1943-51. doi: 10.1104/pp.109.142984. Epub 2009 Sep 29. Plant Physiol. 2009. PMID: 19789289 Free PMC article.
References
- Gesteland R F, Cech T R, Atkins J F. The RNA World. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1999.
- Eddy S R. Curr Opin Genet Dev. 1999;9:695–699. - PubMed
- Eddy S R. Nat Rev Genet. 2001;2:919–929. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases