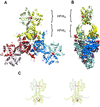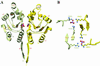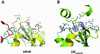Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: Mimicking the product/substrate of the phospho transfer reactions - PubMed (original) (raw)
Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: Mimicking the product/substrate of the phospho transfer reactions
Jose Antonio Márquez et al. Proc Natl Acad Sci U S A. 2002.
Abstract
The histidine containing phospho carrier protein (HPr) kinase/phosphatase is involved in carbon catabolite repression, mainly in Gram-positive bacteria. It is a bifunctional enzyme that phosphorylates Ser-46-HPr in an ATP-dependent reaction and dephosphorylates P-Ser-46-HPr. X-ray analysis of the full-length crystalline enzyme from Staphylococcus xylosus at a resolution of 1.95 A shows the enzyme to consist of two clearly separated domains that are assembled in a hexameric structure resembling a three-bladed propeller. The N-terminal domain has a betaalphabeta fold similar to a segment from enzyme I of the sugar phosphotransferase system and to the uridyl-binding portion of MurF; it is structurally organized in three dimeric modules exposed to form the propeller blades. Two unexpected phosphate ions associated with highly conserved residues were found in the N-terminal dimeric interface. The C-terminal kinase domain is similar to that of the Lactobacillus casei enzyme and is assembled in six copies to form the compact central hub of the propeller. Beyond previously reported similarity with adenylate kinase, we suggest evolutionary relationship with phosphoenolpyruvate carboxykinase. In addition to a phosphate ion in the phosphate-binding loop of the kinase domain, we have identified a second phosphate-binding site that, by comparison with adenylate kinases, we believe accommodates a product/substrate phosphate, normally covalently linked to Ser-46 of HPr. Thus, we propose that our structure represents a product/substrate mimic of the kinase/phosphatase reaction.
Figures
Figure 1
Structure of the full-length HPr protein kinase from Staphylococcus xylosus. (A) Front view of the HPrK hexamer, composed of three structurally identical dimers deeply intertwined. The kinase domains (HPrKC) occupy the central part of the structure whereas the N-terminal domains (HPrKN) extend outwards as pairwise blades in a propeller. Phosphate ions in the interface between N-terminal domains and P-loop regions are depicted as CPK models. (B) Side view of the hexamer. (C) Stereo view of the HPrK dimer representing the asymmetric unit of the crystal.
Figure 2
Multiple sequence alignment of representative members of the HPrK protein family with the secondary structure assignment included (Top), as derived by the program DSSP (47). N-terminal (HPrKN) and C-terminal (HPrKC) domains are indicated. Dashed lines show sequence regions that are not ordered in the crystals (see text). The P-loop region is indicated as a red box. Pink squares denote residues that interact with phosphate ions, and green dots indicate amino acids involved in inter-subunit interactions in the N-terminal (light green) and C-terminal (dark green) domains. In HPrKC, light-pink squares indicate typical P-loop phosphate interactions, and dark-pink squares mark interactions with the second phosphate molecule (see text). Residues K157 and R202 contact both phosphates. Based on structural similarities to NMP kinases and PCK, the region composed of amino acids 263 to 268 (cyan box) is predicted to interact with the adenine moiety of the ATP.
Figure 3
HPrKN and phosphate binding. (A) Ribbon diagram of the dimeric HPrKN, colored as in Fig. 1. The first strand of the C-terminal domain (β6) has been included to illustrate the relative orientation of N-terminal and C-terminal domains (see also Fig. 1_C_). The two phosphate ions, represented in ball and stick, are stabilized by interactions with residues at the subunit interface. Details of these interactions are displayed in B and Fig. 5. (B) Six arginine residues, contributed by both subunits, interact with the phosphate ions to form an almost planar network (bottom view with respect to A).
Figure 4
HPrKC and phosphate binding. (A) Two phosphate ions are bound in the P-loop region, stabilized by invariant residues, most likely involved in catalysis (H136, K157, Ser-153, Glu-159, R202, E200) and/or metal binding (Asp-174/175) (see text). Phosphate 1 occupies the position of the β-phosphate of ATP and is additionally stabilized by water-mediated contacts with R267 from a neighboring subunit. Phosphate 2 (closest to S153) we propose to represent the position of the γ-phosphate of ATP after its transfer to Ser-46 of HPr. Major interactions stabilizing the phosphate ions are shown as blue dashed lines. H136 seems to adopt alternative conformations in the structure, both of which are displayed. (B) P-loop region of HPrKC (yellow) superimposed on the AdK-related UKyeast, bound to two ADP molecules (PDB code 1uky), shown in ball and stick. Electron density corresponding to the two phosphate ions found in HPrKC has been included, matching nearly perfectly the β-phosphate positions of ADP bound to UKyeast. Residues in UKyeast that are structurally equivalent to those in the P-loop of HPrKC are also displayed. K29 and R142 of UKyeast, equivalent to K157 and R202, contact both phosphate ions and are believed to play a role in catalysis through stabilization of transition states. The putative adenine-binding loops are depicted in cyan; the corresponding loop (blue) from neighboring subunit of HPrK is included.
Similar articles
- Crystal structure of HPr kinase/phosphatase from Mycoplasma pneumoniae.
Allen GS, Steinhauer K, Hillen W, Stülke J, Brennan RG. Allen GS, et al. J Mol Biol. 2003 Feb 28;326(4):1203-17. doi: 10.1016/s0022-2836(02)01378-5. J Mol Biol. 2003. PMID: 12589763 - Characterization of an HPr kinase mutant of Staphylococcus xylosus.
Huynh PL, Jankovic I, Schnell NF, Brückner R. Huynh PL, et al. J Bacteriol. 2000 Apr;182(7):1895-902. doi: 10.1128/JB.182.7.1895-1902.2000. J Bacteriol. 2000. PMID: 10714994 Free PMC article. - Phosphorylation of HPr by the bifunctional HPr Kinase/P-ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion.
Dossonnet V, Monedero V, Zagorec M, Galinier A, Pérez-Martínez G, Deutscher J. Dossonnet V, et al. J Bacteriol. 2000 May;182(9):2582-90. doi: 10.1128/JB.182.9.2582-2590.2000. J Bacteriol. 2000. PMID: 10762262 Free PMC article. - HPr kinase/phosphorylase, a Walker motif A-containing bifunctional sensor enzyme controlling catabolite repression in Gram-positive bacteria.
Poncet S, Mijakovic I, Nessler S, Gueguen-Chaignon V, Chaptal V, Galinier A, Boël G, Mazé A, Deutscher J. Poncet S, et al. Biochim Biophys Acta. 2004 Mar 11;1697(1-2):123-35. doi: 10.1016/j.bbapap.2003.11.018. Biochim Biophys Acta. 2004. PMID: 15023355 Review. - The bacterial HPr kinase/phosphorylase: a new type of Ser/Thr kinase as antimicrobial target.
Nessler S. Nessler S. Biochim Biophys Acta. 2005 Dec 30;1754(1-2):126-31. doi: 10.1016/j.bbapap.2005.07.042. Epub 2005 Sep 8. Biochim Biophys Acta. 2005. PMID: 16202671 Review.
Cited by
- X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Fieulaine S, Morera S, Poncet S, Mijakovic I, Galinier A, Janin J, Deutscher J, Nessler S. Fieulaine S, et al. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13437-41. doi: 10.1073/pnas.192368699. Epub 2002 Oct 1. Proc Natl Acad Sci U S A. 2002. PMID: 12359875 Free PMC article. - CcpA-dependent carbon catabolite repression in bacteria.
Warner JB, Lolkema JS. Warner JB, et al. Microbiol Mol Biol Rev. 2003 Dec;67(4):475-90. doi: 10.1128/MMBR.67.4.475-490.2003. Microbiol Mol Biol Rev. 2003. PMID: 14665673 Free PMC article. Review. - In vivo activity of enzymatic and regulatory components of the phosphoenolpyruvate:sugar phosphotransferase system in Mycoplasma pneumoniae.
Halbedel S, Hames C, Stülke J. Halbedel S, et al. J Bacteriol. 2004 Dec;186(23):7936-43. doi: 10.1128/JB.186.23.7936-7943.2004. J Bacteriol. 2004. PMID: 15547265 Free PMC article. - On the emergence of P-Loop NTPase and Rossmann enzymes from a Beta-Alpha-Beta ancestral fragment.
Longo LM, Jabłońska J, Vyas P, Kanade M, Kolodny R, Ben-Tal N, Tawfik DS. Longo LM, et al. Elife. 2020 Dec 9;9:e64415. doi: 10.7554/eLife.64415. Elife. 2020. PMID: 33295875 Free PMC article. - Bacterial tyrosine kinases: evolution, biological function and structural insights.
Grangeasse C, Nessler S, Mijakovic I. Grangeasse C, et al. Philos Trans R Soc Lond B Biol Sci. 2012 Sep 19;367(1602):2640-55. doi: 10.1098/rstb.2011.0424. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22889913 Free PMC article. Review.
References
- Lengeler J W, Vogler A P. FEMS Microbiol Rev. 1989;5:81–92. - PubMed
- Gunnewijk M G, van den Bogaard P T, Veenhoff L M, Heuberger E H, de Vos W M, Kleerebezem M, Kuipers O P, Poolman B. J Mol Microbiol Biotechnol. 2001;3:401–413. - PubMed
- Inada T, Kimata K, Aiba H. Genes Cells. 1996;1:293–301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources