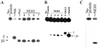Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA - PubMed (original) (raw)
Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA
Tapas K Hazra et al. Proc Natl Acad Sci U S A. 2002.
Abstract
8-oxoguanine (8-oxoG), ring-opened purines (formamidopyrimidines or Fapys), and other oxidized DNA base lesions generated by reactive oxygen species are often mutagenic and toxic, and have been implicated in the etiology of many diseases, including cancer, and in aging. Repair of these lesions in all organisms occurs primarily via the DNA base excision repair pathway, initiated with their excision by DNA glycosylase/AP lyases, which are of two classes. One class utilizes an internal Lys residue as the active site nucleophile, and includes Escherichia coli Nth and both known mammalian DNA glycosylase/AP lyases, namely, OGG1 and NTH1. E. coli MutM and its paralog Nei, which comprise the second class, use N-terminal Pro as the active site. Here, we report the presence of two human orthologs of E. coli mutM nei genes in the human genome database, and characterize one of their products. Based on the substrate preference, we have named it NEH1 (Nei homolog). The 44-kDa, wild-type recombinant NEH1, purified to homogeneity from E. coli, excises Fapys from damaged DNA, and oxidized pyrimidines and 8-oxoG from oligodeoxynucleotides. Inactivation of the enzyme because of either deletion of N-terminal Pro or Histag fusion at the N terminus supports the role of N-terminal Pro as its active site. The tissue-specific levels of NEH1 and OGG1 mRNAs are distinct, and S phase-specific increase in NEH1 at both RNA and protein levels suggests that NEH1 is involved in replication-associated repair of oxidized bases.
Figures
Figure 1
Sequence alignment of hNEH1 and hNEH2 with _E. coli_MutM/Nei. Identical residues are underlined. Box A contains catalytic N-terminal Pro; box B, helix-2-turn-helix motif; box C, Zn-finger motif. Potential nuclear localization signal (NLS; by
psort
) in NEH1 is underlined. N-terminal Met is cleaved after synthesis.
Figure 2
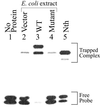
Trapping analysis of recombinant NEH1 in extracts of mutM nei E. coli. Lane 1, no protein; lanes 2–4, trapping assay with lysates (1 μg) of E. coli containing empty vector, WT NEH1, and Pro 1 deletion mutant, after incubation with 0.1 pmol of DHU⋅A-containing duplex oligo; lane 5, trapped complex of purified Nth (5 ng) used as a reference with 0.1 pmol of DHU⋅A. The positions of trapped complexes and free DNA are indicated.
Figure 3
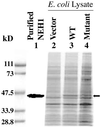
Expression and purification of NEH1. Lysates (15 μg) of_E. coli_ expressing NEH1 were analyzed by SDS/PAGE and Coomassie blue staining. The Bio-Rad protein markers are shown on the left. Purified WT NEH1 (lane 1); lanes 2–4, lysates of_E. coli_ with empty vector, WT, and Pro-1 mutant NEH1 expression plasmid, respectively. The position of NEH1 is indicated by arrow.
Figure 4
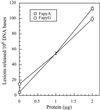
Excision of FapyA (□) and FapyG (○) by hNEH1 from irradiated DNA. Other details are described in_Materials and Methods_.
Figure 5
Substrate specificity and AP lyase activity of purified hNEH1. (A) Purified NEH1 (0.5 pmol) was incubated with 5′32P-labeled DHU-containing duplex oligo (4 pmol) with A, T, G, and C in the complementary strand (lanes 4–7) opposite DHU. Lanes 2 and 3, Incision activity of Nth and Nei, respectively, with DHU⋅A oligo. (B) Incision activity of NEH1 with 8-oxoG⋅A-, 8-oxoG⋅T-, 8-oxoG⋅G-, and 8-oxoG⋅C-containing oligos, respectively (lanes 2–5). Incision activity of 8-oxoG⋅C with hOGG1 and MutM (lanes 6 and 7). Positions of β and βδ-elimination products are indicated. (C) Cleavage of 5-OHU-containing oligo (100 fmol) by NEH1. Lane 1, No enzyme; lane 2, 1 pmol NEH1.
Figure 6
Tissue-specific expression of NEH1. The blots containing 2 μg poly(A)+ RNA purified from various human tissues (CLONTECH) were probed sequentially for Northern analysis with32P-labeled cDNAs of hNEH1 (Upper) and hOGG1 (Lower).
Figure 7
S phase-specific activation of NEH1. (A) Northern analysis of NEH1 mRNA, and (B) Western analysis of NEH1 polypeptide in synchronized MRC5 cells collected at various times after serum addition. The percentage of S phase cells in these different experiments is shown.
Similar articles
- Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions.
Hazra TK, Hill JW, Izumi T, Mitra S. Hazra TK, et al. Prog Nucleic Acid Res Mol Biol. 2001;68:193-205. doi: 10.1016/s0079-6603(01)68100-5. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554297 Review. - Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites.
Bjorâs M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Bjorâs M, et al. EMBO J. 1997 Oct 15;16(20):6314-22. doi: 10.1093/emboj/16.20.6314. EMBO J. 1997. PMID: 9321410 Free PMC article. - Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine.
van der Kemp PA, Thomas D, Barbey R, de Oliveira R, Boiteux S. van der Kemp PA, et al. Proc Natl Acad Sci U S A. 1996 May 28;93(11):5197-202. doi: 10.1073/pnas.93.11.5197. Proc Natl Acad Sci U S A. 1996. PMID: 8643552 Free PMC article. - Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily.
Nash HM, Bruner SD, Schärer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Nash HM, et al. Curr Biol. 1996 Aug 1;6(8):968-80. doi: 10.1016/s0960-9822(02)00641-3. Curr Biol. 1996. PMID: 8805338 - Repair of oxidative DNA damage: mechanisms and functions.
Lu AL, Li X, Gu Y, Wright PM, Chang DY. Lu AL, et al. Cell Biochem Biophys. 2001;35(2):141-70. doi: 10.1385/CBB:35:2:141. Cell Biochem Biophys. 2001. PMID: 11892789 Review.
Cited by
- Oxidized Guanine Base Lesions Function in 8-Oxoguanine DNA Glycosylase-1-mediated Epigenetic Regulation of Nuclear Factor κB-driven Gene Expression.
Pan L, Zhu B, Hao W, Zeng X, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR, Ba X, Boldogh I. Pan L, et al. J Biol Chem. 2016 Dec 2;291(49):25553-25566. doi: 10.1074/jbc.M116.751453. Epub 2016 Oct 18. J Biol Chem. 2016. PMID: 27756845 Free PMC article. - Oxidized base damage and single-strand break repair in mammalian genomes: role of disordered regions and posttranslational modifications in early enzymes.
Hegde ML, Izumi T, Mitra S. Hegde ML, et al. Prog Mol Biol Transl Sci. 2012;110:123-53. doi: 10.1016/B978-0-12-387665-2.00006-7. Prog Mol Biol Transl Sci. 2012. PMID: 22749145 Free PMC article. Review. - Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice.
Møllersen L, Rowe AD, Illuzzi JL, Hildrestrand GA, Gerhold KJ, Tveterås L, Bjølgerud A, Wilson DM 3rd, Bjørås M, Klungland A. Møllersen L, et al. Hum Mol Genet. 2012 Nov 15;21(22):4939-47. doi: 10.1093/hmg/dds337. Epub 2012 Aug 21. Hum Mol Genet. 2012. PMID: 22914735 Free PMC article. - 8-Hydroxyguanine: From its discovery in 1983 to the present status.
Nishimura S. Nishimura S. Proc Jpn Acad Ser B Phys Biol Sci. 2006 May;82(4):127-41. doi: 10.2183/pjab.82.127. Proc Jpn Acad Ser B Phys Biol Sci. 2006. PMID: 25792776 Free PMC article. Review. - Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage.
Liu M, Doublié S, Wallace SS. Liu M, et al. Mutat Res. 2013 Mar-Apr;743-744:4-11. doi: 10.1016/j.mrfmmm.2012.12.003. Epub 2012 Dec 26. Mutat Res. 2013. PMID: 23274422 Free PMC article. Review.
References
- Grisham M B, McCord J M. In: Physiology of Oxygen Radicals. Taylor A E, Matalon S, Ward P A, editors. Baltimore: Waverly Press; 1986. pp. 1–18.
- Gotz M E, Kunig G, Riederer P, Youdim M B. Pharmacol Ther. 1994;63:37–122. - PubMed
- Lovell M A, Xie C, Markesbery W R. Brain Res. 2000;855:116–123. - PubMed
- Breen A P, Murphy J A. Free Rad Biol Med. 1995;18:1033–1077. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA081063/CA/NCI NIH HHS/United States
- R01 CA084461/CA/NCI NIH HHS/United States
- CA81063/CA/NCI NIH HHS/United States
- CA 84461/CA/NCI NIH HHS/United States
- R01 CA090860/CA/NCI NIH HHS/United States
- P30 ES006676/ES/NIEHS NIH HHS/United States
- CA90860/CA/NCI NIH HHS/United States
- ES06676/ES/NIEHS NIH HHS/United States
- R56 CA090860/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials