Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae - PubMed (original) (raw)
Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae
Kai Stefan Dimmer et al. Mol Biol Cell. 2002 Mar.
Abstract
The understanding of the processes underlying organellar function and inheritance requires the identification and characterization of the molecular components involved. We pursued a genomic approach to define the complements of genes required for respiratory growth and inheritance of mitochondria with normal morphology in yeast. With the systematic screening of a deletion mutant library covering the nonessential genes of Saccharomyces cerevisiae the numbers of genes known to be required for respiratory function and establishment of wild-type-like mitochondrial structure have been more than doubled. In addition to the identification of novel components, the systematic screen revealed unprecedented mitochondrial phenotypes that have never been observed by conventional screens. These data provide a comprehensive picture of the cellular processes and molecular components required for mitochondrial function and structure in a simple eukaryotic cell.
Figures
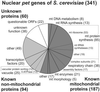
Figure 1
Distribution of functional classes of known and newly identified yeast pet genes using criteria from the Yeast Proteome Database (Costanzo et al., 2001). Numbers indicate the number of pet genes falling into each functional class. An annotated list of all identified pet genes catalogued according to their cellular function can be found in supplemental Table 3. mt, mitochondrial.
Figure 2
Mitochondrial morphology of newly identified mdm mutants. Strains expressing mitochondria-targeted GFP were grown in glucose-containing YPD medium at 30°C to logarithmic growth phase and subjected to fluorescence microscopy. Left, mitochondrial morphology of representative cells; right, overlay with the corresponding phase contrast image. WT, wild-type. Bar, 5 μm.
Similar articles
- Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae.
Westermann B, Neupert W. Westermann B, et al. Yeast. 2000 Nov;16(15):1421-7. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. Yeast. 2000. PMID: 11054823 - Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae.
Altmann K, Westermann B. Altmann K, et al. Mol Biol Cell. 2005 Nov;16(11):5410-7. doi: 10.1091/mbc.e05-07-0678. Epub 2005 Aug 31. Mol Biol Cell. 2005. PMID: 16135527 Free PMC article. - Mitochondrial inheritance in yeast.
Westermann B. Westermann B. Biochim Biophys Acta. 2014 Jul;1837(7):1039-46. doi: 10.1016/j.bbabio.2013.10.005. Epub 2013 Oct 29. Biochim Biophys Acta. 2014. PMID: 24183694 Review. - Molecular machinery of mitochondrial dynamics in yeast.
Merz S, Hammermeister M, Altmann K, Dürr M, Westermann B. Merz S, et al. Biol Chem. 2007 Sep;388(9):917-26. doi: 10.1515/BC.2007.110. Biol Chem. 2007. PMID: 17696775 Review.
Cited by
- Regulating the metabolic flux of pyruvate dehydrogenase bypass to enhance lipid production in Saccharomyces cerevisiae.
Lei C, Guo X, Zhang M, Zhou X, Ding N, Ren J, Liu M, Jia C, Wang Y, Zhao J, Dong Z, Lu D. Lei C, et al. Commun Biol. 2024 Oct 26;7(1):1399. doi: 10.1038/s42003-024-07103-7. Commun Biol. 2024. PMID: 39462103 Free PMC article. - Co-utilization of microalgae and heterotrophic microorganisms improves wastewater treatment efficiency.
Takahashi M, Karitani Y, Yamada R, Matsumoto T, Ogino H. Takahashi M, et al. Appl Microbiol Biotechnol. 2024 Sep 18;108(1):468. doi: 10.1007/s00253-024-13309-w. Appl Microbiol Biotechnol. 2024. PMID: 39292263 Free PMC article. - Drosophila Clueless ribonucleoprotein particles display novel dynamics that rely on the availability of functional protein and polysome equilibrium.
Hwang HJ, Sheard KM, Cox RT. Hwang HJ, et al. bioRxiv [Preprint]. 2024 Aug 22:2024.08.21.609023. doi: 10.1101/2024.08.21.609023. bioRxiv. 2024. PMID: 39229069 Free PMC article. Preprint. - The Mitochondrial Distribution and Morphology Family 33 Gene FgMDM33 Is Involved in Autophagy and Pathogenesis in Fusarium graminearum.
Lv W, Tu Y, Xu T, Zhang Y, Chen J, Yang N, Wang Y. Lv W, et al. J Fungi (Basel). 2024 Aug 16;10(8):579. doi: 10.3390/jof10080579. J Fungi (Basel). 2024. PMID: 39194905 Free PMC article. - The ER-SURF pathway uses ER-mitochondria contact sites for protein targeting to mitochondria.
Koch C, Lenhard S, Räschle M, Prescianotto-Baschong C, Spang A, Herrmann JM. Koch C, et al. EMBO Rep. 2024 Apr;25(4):2071-2096. doi: 10.1038/s44319-024-00113-w. Epub 2024 Apr 2. EMBO Rep. 2024. PMID: 38565738 Free PMC article.
References
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. - PubMed
- Bereiter-Hahn J, Vöth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases