Subcellular localization of the yeast proteome - PubMed (original) (raw)
. 2002 Mar 15;16(6):707-19.
doi: 10.1101/gad.970902.
Seema Agarwal, John A Heyman, Sandra Matson, Matthew Heidtman, Stacy Piccirillo, Lara Umansky, Amar Drawid, Ronald Jansen, Yang Liu, Kei-Hoi Cheung, Perry Miller, Mark Gerstein, G Shirleen Roeder, Michael Snyder
Affiliations
- PMID: 11914276
- PMCID: PMC155358
- DOI: 10.1101/gad.970902
Subcellular localization of the yeast proteome
Anuj Kumar et al. Genes Dev. 2002.
Abstract
Protein localization data are a valuable information resource helpful in elucidating eukaryotic protein function. Here, we report the first proteome-scale analysis of protein localization within any eukaryote. Using directed topoisomerase I-mediated cloning strategies and genome-wide transposon mutagenesis, we have epitope-tagged 60% of the Saccharomyces cerevisiae proteome. By high-throughput immunolocalization of tagged gene products, we have determined the subcellular localization of 2744 yeast proteins. Extrapolating these data through a computational algorithm employing Bayesian formalism, we define the yeast localizome (the subcellular distribution of all 6100 yeast proteins). We estimate the yeast proteome to encompass approximately 5100 soluble proteins and >1000 transmembrane proteins. Our results indicate that 47% of yeast proteins are cytoplasmic, 13% mitochondrial, 13% exocytic (including proteins of the endoplasmic reticulum and secretory vesicles), and 27% nuclear/nucleolar. A subset of nuclear proteins was further analyzed by immunolocalization using surface-spread preparations of meiotic chromosomes. Of these proteins, 38% were found associated with chromosomal DNA. As determined from phenotypic analyses of nuclear proteins, 34% are essential for spore viability--a percentage nearly twice as great as that observed for the proteome as a whole. In total, this study presents experimentally derived localization data for 955 proteins of previously unknown function: nearly half of all functionally uncharacterized proteins in yeast. To facilitate access to these data, we provide a searchable database featuring 2900 fluorescent micrographs at http://ygac.med.yale.edu.
Figures
Figure 1
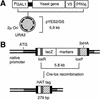
Genome-wide epitope-tagging strategies. (A) Yeast ORFs were amplified by PCR and cloned by topoisomerase I-mediated ligation into the yeast expression vector pYES2/GS. The pYES2/GS vector carries the yeast 2μ origin of replication for maintenance of high copy number. Yeast genes were inserted into pYES2 such that they are under transcriptional control of the GAL1 promoter and fused at their 3′ ends to sequence encoding the V5-epitope and polyhistidine tag (HIS)6. By galactose induction in yeast, cloned genes were overexpressed as V5-tagged proteins for subsequent immunolocalization with α-V5 antibodies (in 96-well formats). (B) Modified bacterial transposons were used to randomly tag yeast genes at their native genomic loci with sequence encoding three copies of the viral haemagglutinin epitope (3×HA epitope). The transposon carries a promoterless and 5′-truncated lacZ reporter enabling selection of in-frame insertions by β-galactosidase assay. In-frame insertions were subsequently modified in yeast by Cre-lox recombination, such that the majority of the transposon sequence was excised. The remaining HA-epitope insertion element (HAT tag) encodes no stop codons in the specified reading frame. The indicated 279-bp HAT-tag insertion includes a 5-bp duplication in target site sequence associated with Tn_3_ transposition. HAT-tagged proteins were immunolocalized with monoclonal α-HA antibodies in a 96-well format.
Figure 2
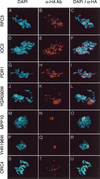
Immunolocalization of epitope-tagged proteins. (A_–_E) Vegetative cells containing HAT-tagged proteins were stained with the DNA-binding dye 4‘,6-diamidino-2-phenylindole (DAPI; left image) and monoclonal antibody against HA (center). Per row, the DAPI-stained and α-HA-stained images are shown merged in the rightmost panel. Typical nucleolar staining patterns can be seen in strains containing HAT-tagged alleles of the rRNA-binding proteins Net1p (A) and Sik1p (E). Staining of the cell neck is evident in cells containing HAT-tagged Hsl1p (B). HAT-tagging of the vacuolar ATPase Vma6p is shown in row C. Staining of the cell periphery can be seen upon HAT-tagging of the cell surface glycoprotein Gas1p (D). (F_–_J) Vegetative cells carrying V5-tagged proteins were stained with monoclonal antibody directed against the V5 epitope (center). Corresponding DAPI-stained images and merged images are shown to the left and right, respectively. Nucleolar staining is apparent in cells carrying V5-tagged Nop13p (F). Note, however, that V5-tagging and mild overexpression of SIK1 (J) results in a nuclear staining pattern, as opposed to the nucleolar pattern evident upon HAT-tagging of this same gene (E). Mitochondrial staining (G) can be seen in cells carrying a tagged allele of YMR293C; overlap between DAPI- and α-V5 staining is shown in the merged image. V5-tagged Gpi12p localizes to the endoplasmic reticulum (H), visible as an area of strong staining around the nuclear rim. A patchy pattern of cytoplasmic staining can be seen in cells carrying V5-tagged Bzz1p (I). Bar, 2 μm.
Figure 3
Subcellular compartmentalization of the yeast proteome. (A) Cellular compartments are as follows: cytoplasmic (Cyt.), nuclear (Nuc.), mitochondrial (Mit.), and exocytic (Exo.). The membrane fraction of each compartment is indicated in stripes. The percentage of the yeast proteome contained within the respective membrane and soluble fractions of each compartment is indicated outside the chart; the total percentage of the proteome contained within each of the four main compartments is indicated inside the chart. Plasma membrane proteins are included in the cytoplasmic compartment for purposes of this analysis. (B) The corresponding protein population of each cellular compartment and membrane/soluble subfraction is indicated.
Figure 4
Immunolocalization of nuclear proteins on surface-spread meiotic chromosomes. Meiotic chromosomes were surface spread and stained with the DNA-binding dye DAPI (left) and monoclonal anti-HA antibodies (center). Corresponding merged images are shown to the right. A general pattern of chromosomal binding can be seen from immunofluorescence analysis of cells containing HAT-tagged alleles of RFC3 (A_–_C), IOC2 (D_–_F), and PDR1 (G_–_I). Nine proteins localized predominantly to the nucleolus; typical nucleolar staining patterns are shown here in cells containing HAT-tagged alleles of YGR090W (J_–_L), MPP10 (M_–_O), and YHR196W (P_–_R). Specific binding to telomeric DNA can be seen upon HAT-tagging and immunolocalization of the origin recognition complex subunit Orc4p (S_–_U). Bar, 1 μm.
Figure 5
Chromosomal localization and phenotypic analysis of nuclear proteins. Chromosomal localization indicates a general pattern of chromosomal binding, typically with >40 staining foci per nucleus. Strains disrupted for each gene were assayed for spore viability or growth defects; observed disruption mutants are categorized as viable, inviable, or slow-growth, accordingly.
Figure 6
Prevalent functions associated with cellular compartments in yeast. Functional categorizations (compiled from published literature) were extracted from the MIPS CYG database for all proteins experimentally localized in this study. In total, functions were available for 1789 proteins; the number of functionally categorized proteins localized to each of the indicated cellular compartments is shown. Mixed localizations are also represented: 165 functionally characterized proteins were colocalized to the cytoplasm and nucleus; similarly, 61 such proteins were colocalized to the cytoplasm and endoplasmic reticulum (ER). Functions were tallied for all proteins within a given cellular compartment. The most frequently occurring functions per compartment are shown boxed. Multiple functions may be associated with a single protein. Therefore, the listed percentage following each function refers to the fraction of proteins within each compartment associated with that particular cellular process, and the sum total of these percentages within a given compartment will not equal 100%.
Similar articles
- The TRIPLES database: a community resource for yeast molecular biology.
Kumar A, Cheung KH, Tosches N, Masiar P, Liu Y, Miller P, Snyder M. Kumar A, et al. Nucleic Acids Res. 2002 Jan 1;30(1):73-5. doi: 10.1093/nar/30.1.73. Nucleic Acids Res. 2002. PMID: 11752258 Free PMC article. - A Bayesian system integrating expression data with sequence patterns for localizing proteins: comprehensive application to the yeast genome.
Drawid A, Gerstein M. Drawid A, et al. J Mol Biol. 2000 Aug 25;301(4):1059-75. doi: 10.1006/jmbi.2000.3968. J Mol Biol. 2000. PMID: 10966805 - CYCLoPs: A Comprehensive Database Constructed from Automated Analysis of Protein Abundance and Subcellular Localization Patterns in Saccharomyces cerevisiae.
Koh JL, Chong YT, Friesen H, Moses A, Boone C, Andrews BJ, Moffat J. Koh JL, et al. G3 (Bethesda). 2015 Apr 15;5(6):1223-32. doi: 10.1534/g3.115.017830. G3 (Bethesda). 2015. PMID: 26048563 Free PMC article. - Yeast as a model eukaryote in toxinology: a functional genomics approach to studying the molecular basis of action of pharmacologically active molecules.
Mattiazzi M, Petrovič U, Križaj I. Mattiazzi M, et al. Toxicon. 2012 Sep 15;60(4):558-71. doi: 10.1016/j.toxicon.2012.03.014. Epub 2012 Mar 21. Toxicon. 2012. PMID: 22465496 Review. - Protein pI and Intracellular Localization.
Tokmakov AA, Kurotani A, Sato KI. Tokmakov AA, et al. Front Mol Biosci. 2021 Nov 29;8:775736. doi: 10.3389/fmolb.2021.775736. eCollection 2021. Front Mol Biosci. 2021. PMID: 34912847 Free PMC article. Review.
Cited by
- Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols.
Avalos JL, Fink GR, Stephanopoulos G. Avalos JL, et al. Nat Biotechnol. 2013 Apr;31(4):335-41. doi: 10.1038/nbt.2509. Epub 2013 Feb 17. Nat Biotechnol. 2013. PMID: 23417095 Free PMC article. - Detection of protein-protein interactions at the septin collar in Saccharomyces cerevisiae using a tripartite split-GFP system.
Finnigan GC, Duvalyan A, Liao EN, Sargsyan A, Thorner J. Finnigan GC, et al. Mol Biol Cell. 2016 Sep 1;27(17):2708-25. doi: 10.1091/mbc.E16-05-0337. Epub 2016 Jul 6. Mol Biol Cell. 2016. PMID: 27385335 Free PMC article. - Assembly of the yeast exoribonuclease Rrp6 with its associated cofactor Rrp47 occurs in the nucleus and is critical for the controlled expression of Rrp47.
Feigenbutz M, Jones R, Besong TM, Harding SE, Mitchell P. Feigenbutz M, et al. J Biol Chem. 2013 May 31;288(22):15959-70. doi: 10.1074/jbc.M112.445759. Epub 2013 Apr 11. J Biol Chem. 2013. PMID: 23580640 Free PMC article. - Improved tagging strategy for protein identification in mammalian cells.
Bialkowska A, Zhang XY, Reiser J. Bialkowska A, et al. BMC Genomics. 2005 Sep 4;6:113. doi: 10.1186/1471-2164-6-113. BMC Genomics. 2005. PMID: 16138932 Free PMC article. - Protein interaction networks by proteome peptide scanning.
Landgraf C, Panni S, Montecchi-Palazzi L, Castagnoli L, Schneider-Mergener J, Volkmer-Engert R, Cesareni G. Landgraf C, et al. PLoS Biol. 2004 Jan;2(1):E14. doi: 10.1371/journal.pbio.0020014. Epub 2004 Jan 20. PLoS Biol. 2004. PMID: 14737190 Free PMC article.
References
- Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. - PubMed
- Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. - PubMed
- Burley SK. An overview of structural genomics. Nat Struct Biol. 2000;Suppl:932–934. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous