Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities - PubMed (original) (raw)
Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities
Andreas Teske et al. Appl Environ Microbiol. 2002 Apr.
Abstract
Microbial communities in hydrothermally active sediments of the Guaymas Basin (Gulf of California, Mexico) were studied by using 16S rRNA sequencing and carbon isotopic analysis of archaeal and bacterial lipids. The Guaymas sediments harbored uncultured euryarchaeota of two distinct phylogenetic lineages within the anaerobic methane oxidation 1 (ANME-1) group, ANME-1a and ANME-1b, and of the ANME-2c lineage within the Methanosarcinales, both previously assigned to the methanotrophic archaea. The archaeal lipids in the Guaymas Basin sediments included archaeol, diagnostic for nonthermophilic euryarchaeota, and sn-2-hydroxyarchaeol, with the latter compound being particularly abundant in cultured members of the Methanosarcinales. The concentrations of these compounds were among the highest observed so far in studies of methane seep environments. The delta-(13)C values of these lipids (delta-(13)C = -89 to -58 per thousand) indicate an origin from anaerobic methanotrophic archaea. This molecular-isotopic signature was found not only in samples that yielded predominantly ANME-2 clones but also in samples that yielded exclusively ANME-1 clones. ANME-1 archaea therefore remain strong candidates for mediation of the anaerobic oxidation of methane. Based on 16S rRNA data, the Guaymas sediments harbor phylogenetically diverse bacterial populations, which show considerable overlap with bacterial populations of geothermal habitats and natural or anthropogenic hydrocarbon-rich sites. Consistent with earlier observations, our combined evidence from bacterial phylogeny and molecular-isotopic data indicates an important role of some novel deeply branching bacteria in anaerobic methanotrophy. Anaerobic methane oxidation likely represents a significant and widely occurring process in the trophic ecology of methane-rich hydrothermal vents. This study stresses a high diversity among communities capable of anaerobic oxidation of methane.
Figures
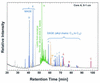
FIG. 1.
Reconstructed ion chromatogram of alcohol fraction from core A sample, 0 to 1 cm. Colors of peaks designate their biological sources: red, methanotrophic archaea; blue, presumed bacterial members of methanotrophic community; orange, aerobic bacteria; green, eucaryotes, e.g., marine algae. Refer to Table 1 for further details.
FIG. 2.
Distance tree of archaeal clones from Guaymas sediment cores A, C, and T, based on near-complete 16S rDNA sequences. Bootstrap values (in percent) are based on 1,000 replicates each (distance and parsimony) and are shown for branches with more than 50% bootstrap support. The sequence for Methanohalophilus zhilinae was obtained from the ARB sequence server (version May 2001). Crenarchaeotal sequences SB-pc17-1A10 and Eel-36a2H12 were provided by Victoria Orphan. For the major phylogenetic divisions, the numbers of core A, core C, and core T clones are given in that order. For multiple near-identical clones (>99% identity), the number of multiples is given after the GenBank number of a representative clone. Some sequence types occurred in several sediment layers: Guaymas clone A1 R010 was found four times in the 0- to 1-cm layer of core A and two times in the 1- to 2-cm layer of core A. Guaymas clone A2 R045 was found 15 times in the 0- to 1-cm layer of core A and 2 times in the 1- to 2-cm layer of core A. Guaymas clone T R022 was found one time in the 0- to 1-cm layer of core C and one time in core T.
FIG. 3.
Distance tree of proteobacterial clones from Guaymas sediment cores A and C, based on near-complete 16S rDNA sequences. Bootstrap values (in percent) are based on 1,000 replicates each (distance and parsimony) and are shown for branches with more than 50% bootstrap support. For the major phylogenetic divisions, the numbers of core A and core C clones are given in that order. For multiple near-identical clones (>99% identity), the number of multiples is given after the GenBank number of a representative clone. Some sequence types occurred in several sediment layers: Guaymas clone A1 B030 was found three times in the 0- to 1-cm layer of core A and one time in the 1- to 2-cm layer of core A, Guaymas clone A2 B043 was found one time in the 0- to 1-cm layer of core A and five times in the 1- to 2-cm layer of core A, and Guaymas clone A2 B004 was found three times in the 0- to 1-cm layer of core A and three times in the 1- to 2-cm layer of core A.
FIG. 4.
Distance tree of bacterial clones, excluding proteobacteria, from Guaymas sediment cores A and C, based on near-complete 16S rDNA sequences. Bootstrap values (in percent) are based on 1,000 replicates each (distance and parsimony) and are shown for branches with more than 50% bootstrap support. For the major phylogenetic divisions, the numbers of core A and core C clones are given in that order. For multiple near-identical clones (>99% identity), the number of multiples is given after the GenBank number of a representative clone. Some sequence types occurred in several sediment layers: Guaymas clone A1 B007 was found two times in the 0- to 1-cm layer and one time in the 1- to 2-cm layer of core A, Guaymas clone A2 B031 was found two times in the 0- to 1-cm layer and one time in the 1- to 2-cm layer of core A, Guaymas clone A1 B020 was found three times in the 0- to 1-cm layer and two times in the 1- to 2-cm layer of core A, and Guaymas clone A2 B002 was found one time in the 0- to 1-cm layer and two times in the 1- to 2-cm layer of core A.
Similar articles
- Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments.
Biddle JF, Cardman Z, Mendlovitz H, Albert DB, Lloyd KG, Boetius A, Teske A. Biddle JF, et al. ISME J. 2012 May;6(5):1018-31. doi: 10.1038/ismej.2011.164. Epub 2011 Nov 17. ISME J. 2012. PMID: 22094346 Free PMC article. - Archaeal and anaerobic methane oxidizer communities in the Sonora Margin cold seeps, Guaymas Basin (Gulf of California).
Vigneron A, Cruaud P, Pignet P, Caprais JC, Cambon-Bonavita MA, Godfroy A, Toffin L. Vigneron A, et al. ISME J. 2013 Aug;7(8):1595-608. doi: 10.1038/ismej.2013.18. Epub 2013 Feb 28. ISME J. 2013. PMID: 23446836 Free PMC article. - Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments.
Orphan VJ, Hinrichs KU, Ussler W 3rd, Paull CK, Taylor LT, Sylva SP, Hayes JM, Delong EF. Orphan VJ, et al. Appl Environ Microbiol. 2001 Apr;67(4):1922-34. doi: 10.1128/AEM.67.4.1922-1934.2001. Appl Environ Microbiol. 2001. PMID: 11282650 Free PMC article. - Physiology and Distribution of Archaeal Methanotrophs That Couple Anaerobic Oxidation of Methane with Sulfate Reduction.
Bhattarai S, Cassarini C, Lens PNL. Bhattarai S, et al. Microbiol Mol Biol Rev. 2019 Jul 31;83(3):e00074-18. doi: 10.1128/MMBR.00074-18. Print 2019 Aug 21. Microbiol Mol Biol Rev. 2019. PMID: 31366606 Free PMC article. Review. - Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review.
Valentine DL. Valentine DL. Antonie Van Leeuwenhoek. 2002 Aug;81(1-4):271-82. doi: 10.1023/a:1020587206351. Antonie Van Leeuwenhoek. 2002. PMID: 12448726 Review.
Cited by
- Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center.
Anantharaman K, Breier JA, Dick GJ. Anantharaman K, et al. ISME J. 2016 Jan;10(1):225-39. doi: 10.1038/ismej.2015.81. Epub 2015 Jun 5. ISME J. 2016. PMID: 26046257 Free PMC article. - Archaeal community structures in the solfataric acidic hot springs with different temperatures and elemental compositions.
Satoh T, Watanabe K, Yamamoto H, Yamamoto S, Kurosawa N. Satoh T, et al. Archaea. 2013;2013:723871. doi: 10.1155/2013/723871. Epub 2013 Apr 22. Archaea. 2013. PMID: 23710131 Free PMC article. - First cultivation and ecological investigation of a bacterium affiliated with the candidate phylum OP5 from hot springs.
Mori K, Sunamura M, Yanagawa K, Ishibashi J, Miyoshi Y, Iino T, Suzuki K, Urabe T. Mori K, et al. Appl Environ Microbiol. 2008 Oct;74(20):6223-9. doi: 10.1128/AEM.01351-08. Epub 2008 Sep 5. Appl Environ Microbiol. 2008. PMID: 18776034 Free PMC article. - Microbial ecology and biogeochemistry of hypersaline sediments in Orca Basin.
Nigro LM, Elling FJ, Hinrichs KU, Joye SB, Teske A. Nigro LM, et al. PLoS One. 2020 Apr 21;15(4):e0231676. doi: 10.1371/journal.pone.0231676. eCollection 2020. PLoS One. 2020. PMID: 32315331 Free PMC article. - Influence of Environmental Drivers and Potential Interactions on the Distribution of Microbial Communities From Three Permanently Stratified Antarctic Lakes.
Li W, Morgan-Kiss RM. Li W, et al. Front Microbiol. 2019 May 15;10:1067. doi: 10.3389/fmicb.2019.01067. eCollection 2019. Front Microbiol. 2019. PMID: 31156585 Free PMC article.
References
- Angert, E. R., D. E. Northup, A.-L. Reysenbach, A. S. Peek, B. M. Goebel, and N. R. Pace. 1998. Molecular phylogenetic analysis of a bacterial community in Sulphur River, Parker Cave, Kentucky. Am. Mineralogist 83:1583-1592.
- Atkins, M. S., A. P. Teske, and O. R. Anderson. 2000. A survey of flagellate diversity at four deep-sea hydrothermal vents in the Eastern Pacific Ocean using structural and molecular approaches. J. Eukaryot. Microbiol. 47:400-411. - PubMed
- Bazylinski, D. A., J. W. Farrington, and H. W. Jannasch. 1988. Hydrocarbons in surface sediments from a Guaymas Basin hydrothermal vent site. Org. Geochem. 12:547-558.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases