An efficient algorithm for large-scale detection of protein families - PubMed (original) (raw)
An efficient algorithm for large-scale detection of protein families
A J Enright et al. Nucleic Acids Res. 2002.
Abstract
Detection of protein families in large databases is one of the principal research objectives in structural and functional genomics. Protein family classification can significantly contribute to the delineation of functional diversity of homologous proteins, the prediction of function based on domain architecture or the presence of sequence motifs as well as comparative genomics, providing valuable evolutionary insights. We present a novel approach called TRIBE-MCL for rapid and accurate clustering of protein sequences into families. The method relies on the Markov cluster (MCL) algorithm for the assignment of proteins into families based on precomputed sequence similarity information. This novel approach does not suffer from the problems that normally hinder other protein sequence clustering algorithms, such as the presence of multi-domain proteins, promiscuous domains and fragmented proteins. The method has been rigorously tested and validated on a number of very large databases, including SwissProt, InterPro, SCOP and the draft human genome. Our results indicate that the method is ideally suited to the rapid and accurate detection of protein families on a large scale. The method has been used to detect and categorise protein families within the draft human genome and the resulting families have been used to annotate a large proportion of human proteins.
Figures
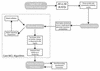
Figure 1
Flowchart of the TRIBE-MCL algorithm.
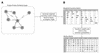
Figure 2
(A) Example of a protein–protein similarity graph for seven proteins (A–F), circles represent proteins (nodes) and lines (edges) represent detected BLASTp similarities with _E_-values (also shown). (B) Weighted transition matrix and associated column stochastic Markov matrix for the seven proteins shown in (A). For explanations, please see text.
Figure 3
Graph representing the largest interconnected group of protein families from the SwissProt protein database (237 protein families, 21 727 sequences in total). Circles represent protein families, with associated family Ids and annotations (where known). Edges show BLAST similarities between families. Circles are coloured according to the GeneOntology (GO) (52) functional class assignments (where available). This graph was generated using the Bio-Layout graph layout algorithm (41).
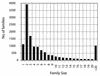
Figure 4
Distribution of protein family sizes within the human genome. The _x_-axis represents family size and the _y_-axis (bars) indicates the number of paralogous protein families.
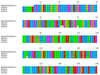
Figure 5
Protein sequence alignment of the eukaryotic TFIIB family of proteins detected using TRIBE-MCL, including three members from SwissProt (accession numbers given) and the human TFIIB (51).
References
- Tsoka S. and Ouzounis,C.A. (2000) Recent developments and future directions in computational genomics. FEBS Lett., 480, 42–48. - PubMed
- Eisenberg D., Marcotte,E.M., Xenarios,I. and Yeates,T.O. (2000) Protein function in the post-genomic era. Nature, 405, 823–826. - PubMed
- Bork P., Dandekar,T., Diaz-Lazcoz,Y., Eisenhaber,F., Huynen,M. and Yuan,Y. (1998) Predicting function: from genes to genomes and back. J. Mol. Biol., 283, 707–725. - PubMed
- Hegyi H. and Gerstein,M. (1999) The relationship between protein structure and function: a comprehensive survey with application to the yeast genome. J. Mol. Biol., 288, 147–164. - PubMed