Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling - PubMed (original) (raw)
Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling
Angela G King et al. Proc Natl Acad Sci U S A. 2002.
Abstract
The most immature lymphoid-committed progenitors in both the bone marrow (common lymphoid progenitor) and thymus (proT1) maintain a latent granulocyte/macrophage (G/M) differentiation potential that can be initiated by signals emanating from exogenously expressed IL-2 receptors. In this study, we investigate at which developmental stage thymocytes lose this G/M differentiation potential. We demonstrate that the next maturational stage after proT1 cells (proT2), but not preT (TN3) cells, can convert cell fate from lymphoid to myeloid in response to ectopic IL-2 receptor signaling in human IL-2Rbeta transgenic mice. It is significant that approximately 10% of clonogenic G/M colonies derived from proT cells of IL-2Rbeta transgenic mice have DJ rearrangement specifically at the Dbeta1 but not Dbeta2 segment in the TCRbeta locus. No TCR gene rearrangement is observed in G/M cells from nontransgenic mice, suggesting that the G/M cells we observe in this system were truly lymphoid-committed before stimulation with IL-2. In addition, Dbeta1 and Dbeta2 DJ rearrangement of the TCRbeta gene may be differentially regulated and thus serve as markers for distinct proT cell maturational stages.
Figures
Figure 1
Schematic diagram of early T cell development. Immigrant cells into the thymus from the bone marrow remain to be identified. HSC, hematopoietic stem cell.
Figure 2
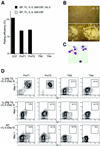
Expression of human IL-2Rβ transgene in CD3−CD4−/loCD8− TN thymocytes. (A) Sorting gates of subpopulations of CD3−CD4−/loCD8− TN thymocytes in this study. (B) Expression of human IL-2Rβ in CD3−CD4−/loCD8− TN thymocytes from IL-2Rβ transgenic mice (filled histogram). Background staining is also shown as a negative control (open histogram). (C Left) 2.5 × 104 proT1 (Upper) and proT2 cells (Lower) from C57BL/Ka-Thy1.1 (Ly5.2) were intravenously injected into 400 rad-irradiated RAG2−/− (Ly5.1) mice. Four weeks after injection, donor-derived cells (Ly5.2+ cells) in the spleen were analyzed by a flow cytometer. (Right) In in vitro stromal cell culture, 40 proT cells were cultured in 96-well plates on an OP9 cell layer in the presence of SIF, FL, IL-7, and human IL-2 for 7 days. Cells in positive wells were pooled and analyzed on a flow cytometer.
Figure 3
A latent myeloid differentiation potential of CD3−CD4−/loCD8− TN thymocytes. (A) G/M colony formation of CD3−CD4−/loCD8− TN thymocytes derived from IL-2Rβ transgenic mice. Two hundred double-sorted cells were cultured in methylcellulose medium for 5–7 days in the presence of cytokines indicated in the figure. (B) Morphology of colonies from IL-2Rβ transgenic proT2 cells after the culture under the condition indicated in A. Only when IL-2 was added did we observe colony formation. (C) Cytospin of G/M colony-forming cells derived from IL-2Rβ transgenic proT2 cells. All colonies we confirmed by cytospin analysis were composed solely of G/M cells. (D) Stromal cell culture of CD3−CD4−/loCD8− TN thymocytes from IL-2Rβ transgenic mice. One hundred double-sorted cells were cultured on OP9 stromal cell layers in the presence of SlF, FL, IL-3, and GM-CSF. Human IL-2 was also added on either Day 0 or Day 4 of the culture as indicated in the figure. Readout populations were analyzed by a flow cytometer after 6 days of the culture. WT, wild type.
Figure 4
Rearrangement of the TCRβ gene in G/M cells derived from IL-2Rβ transgenic proT cells. (A) Schematic diagram of the TCRβ gene locus. Relative position of PCR primers used in this study is shown by arrows. (B) Rearrangement of the TCRβ gene in proT1 and proT2 populations. Genomic DNA was extracted from 5 × 103 double-sorted proT1 and proT2 cells. Dominant germ-line bands were excluded after the first PCR as described in Methods. After nested PCR, amplified products were subjected to 1.2% agarose gel electrophoresis and visualized under UV light after ethydium bromide staining. Spleen T cells (1 × 103) were used as a control. After purification from agarose gel, amplified DNA from the first PCR was serially diluted as indicated in the figure and used as the template for the second PCR. (C) Dβ1Jβ1 (Top) and Dβ2Jβ2 (Bottom) rearrangement was examined in G/M colony-forming cells derived from IL-2Rβ transgenic proT cells. Data shown are representative of five independent colonies derived from proT1 cells of IL-2Rβ transgenic mice. Because the sizes of Dβ1Jβ1.1 and Dβ1Jβ1.2 were too close to be distinguished, these two are denoted by Jβ1.1/1.2 in this study. No Dβ2Jβ2 rearrangement was observed in any of the G/M colonies analyzed. (D) TCRβ gene rearrangement in polyclonal cell populations. Double-sorted IL-2Rβ transgenic proT cells (1 × 105; both proT1 and proT2 cells) were cultured on OP9 stromal cell layers in the presence of SlF, FL, IL-3, GM-CSF, and human IL-2. After 6 days of culture, Gr-1+Mac-1+ G/M cells were sorted and analyzed for TCRβ gene rearrangement by PCR (lane 3). Gr-1+Mac-1+ G/M cells from wild-type (WT) bone marrow were also used in this analysis (lane 4). Amplified products were separated on 1.2% agarose gel and transferred to a nylon membrane. Amplified bands were detected by hybridization with 32P-labeled Dβ1.1 int and Jβ1.7 oligos followed by autoradiography. Genomic DNA from wild-type spleen T cells (lane 2) and RAG2−/− bone marrow cells (lane 1) were used as positive and negative controls, respectively. The band marked with an asterisk (*) is a PCR artifact that is also seen in RAG2−/− cells.
Similar articles
- Enforced granulocyte/macrophage colony-stimulating factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion.
Iwasaki-Arai J, Iwasaki H, Miyamoto T, Watanabe S, Akashi K. Iwasaki-Arai J, et al. J Exp Med. 2003 May 19;197(10):1311-22. doi: 10.1084/jem.20021843. J Exp Med. 2003. PMID: 12756267 Free PMC article. - Antagonistic effect of CCAAT enhancer-binding protein-alpha and Pax5 in myeloid or lymphoid lineage choice in common lymphoid progenitors.
Hsu CL, King-Fleischman AG, Lai AY, Matsumoto Y, Weissman IL, Kondo M. Hsu CL, et al. Proc Natl Acad Sci U S A. 2006 Jan 17;103(3):672-7. doi: 10.1073/pnas.0510304103. Epub 2006 Jan 9. Proc Natl Acad Sci U S A. 2006. PMID: 16407117 Free PMC article. - Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines.
Kondo M, Scherer DC, Miyamoto T, King AG, Akashi K, Sugamura K, Weissman IL. Kondo M, et al. Nature. 2000 Sep 21;407(6802):383-6. doi: 10.1038/35030112. Nature. 2000. PMID: 11014194 - Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells.
Drexler HG. Drexler HG. Leukemia. 1996 Apr;10(4):588-99. Leukemia. 1996. PMID: 8618433 Review. - Early T lymphocyte progenitors.
Shortman K, Wu L. Shortman K, et al. Annu Rev Immunol. 1996;14:29-47. doi: 10.1146/annurev.immunol.14.1.29. Annu Rev Immunol. 1996. PMID: 8717506 Review.
Cited by
- Tumor Arrests DN2 to DN3 Pro T Cell Transition and Promotes Its Conversion to Thymic Dendritic Cells by Reciprocally Regulating Notch1 and Ikaros Signaling.
Guha I, Bhuniya A, Shukla D, Patidar A, Nandi P, Saha A, Dasgupta S, Ganguly N, Ghosh S, Nair A, Majumdar S, Saha B, Storkus WJ, Baral R, Bose A. Guha I, et al. Front Immunol. 2020 Jun 5;11:898. doi: 10.3389/fimmu.2020.00898. eCollection 2020. Front Immunol. 2020. PMID: 32582141 Free PMC article. - Progenitor migration to the thymus and T cell lineage commitment.
Sambandam A, Bell JJ, Schwarz BA, Zediak VP, Chi AW, Zlotoff DA, Krishnamoorthy SL, Burg JM, Bhandoola A. Sambandam A, et al. Immunol Res. 2008;42(1-3):65-74. doi: 10.1007/s12026-008-8035-z. Immunol Res. 2008. PMID: 18827982 Review. - Gene targeting RhoA reveals its essential role in coordinating mitochondrial function and thymocyte development.
Zhang S, Konstantinidis DG, Yang JQ, Mizukawa B, Kalim K, Lang RA, Kalfa TA, Zheng Y, Guo F. Zhang S, et al. J Immunol. 2014 Dec 15;193(12):5973-82. doi: 10.4049/jimmunol.1400839. Epub 2014 Nov 14. J Immunol. 2014. PMID: 25398325 Free PMC article. - Lineage promiscuous expression of transcription factors in normal hematopoiesis.
Miyamoto T, Akashi K. Miyamoto T, et al. Int J Hematol. 2005 Jun;81(5):361-7. doi: 10.1532/ijh97.05003. Int J Hematol. 2005. PMID: 16158815 Review. - A developing picture of lymphopoiesis in bone marrow.
Hirose J, Kouro T, Igarashi H, Yokota T, Sakaguchi N, Kincade PW. Hirose J, et al. Immunol Rev. 2002 Nov;189:28-40. doi: 10.1034/j.1600-065x.2002.18904.x. Immunol Rev. 2002. PMID: 12445263 Free PMC article. Review.
References
- Adkins B, Mueller C, Okada C Y, Reichert R A, Weissman I L, Spangrude G J. Annu Rev Immunol. 1987;5:325–365. - PubMed
- Shortman K, Wu L. Annu Rev Immunol. 1996;14:29–47. - PubMed
- Michie A M, Carlyle J R, Zuniga-Pflucker J C. J Immunol. 1998;160:1735–1741. - PubMed
- Godfrey D I, Kennedy J, Suda T, Zlotnik A. J Immunol. 1993;150:4244–4252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI 47458/AI/NIAID NIH HHS/United States
- T32 CA009302/CA/NCI NIH HHS/United States
- CA 42551/CA/NCI NIH HHS/United States
- AI 47457/AI/NIAID NIH HHS/United States
- R01 AI047458/AI/NIAID NIH HHS/United States
- CA 09302/CA/NCI NIH HHS/United States
- R01 AI047457/AI/NIAID NIH HHS/United States
- R21 AI047458/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical