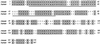Function of quaking in myelination: regulation of alternative splicing - PubMed (original) (raw)
Function of quaking in myelination: regulation of alternative splicing
Jiang I Wu et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Proteomic diversity is frequently achieved by alternative RNA-splicing events that can be fine-tuned in tissue-specific and developmentally regulated ways. Understanding this type of genetic regulation is compelling because of the extensive complexity of alternative splicing found in the nervous system. quaking (qk), one of the classical mouse dysmyelination mutants, is defective for the expression of myelin-associated glycoprotein (MAG), and the misregulation of MAG pre-mRNA alternative splicing is implicated as a causal factor. The qk locus encodes several RNA-binding proteins with heterogeneous nuclear ribonucleoprotein K-type homology, a characteristic of several known alternative splicing regulators. Here we test the nuclear-localized qk isoform (QKI-5) for its ability to regulate alternative splicing of MAG pre-mRNA in transient coexpression assays. QKI-5 exhibits properties of a negative regulator of MAG exon 12 alternative splicing. An intronic sequence element required for the repressive function and binding of QKI-5 is also identified. Direct evidence for irregularities in alternative splicing of MAG and other myelin protein transcripts in the qk mouse is demonstrated.
Figures
Figure 1
Abnormal myelin gene alternative splicing in the qk v mutant. (Upper Right) RT-PCR of MAG, PLP, and MBP messages from normal (+/qk v) and mutant (qk v/qk v) brain RNA from P14 and adult mice. (Upper Left) Diagram of MAG, PLP, and MBP alternative splicing pattern. The arrows represent the primers. The content of each PCR product is indicated next to the band. (Lower) Graphical comparison of the percentage of exon selection.
Figure 2
The repression of MAG exon 12 inclusion by QKI-5. (A) Schematic drawing of the MAG minigene construct. A 1.1-kb genomic DNA fragment around MAG exon 12 is cloned downstream of an HIV tat exon (gray box) in the vector pSPL3. SV40P, simian virus 40 promoter. The numbers above refer to the size (bp) of corresponding exon or intron. The arrows represent the primers used in the RT-PCR in B. The polypyrimidine (PY) tract of exon 12 is indicated by the small black box. In the pMAGPY construct, it was replaced by the PY of exon 11 (from CGTTTCCTTCTTCAAT to ATGTTTCATGCCTCCCTGC). (B) Representative RT-PCR results of RNA from COS7 cells cotransfected with 0.5 μg of MAG minigene and indicated amounts of QKI expression plasmid. The vector pcDNA3 was used to maintain the total level of transfected DNA and as a negative control. The intensity of the bands was measured with a PhosphorImager, and the percentage of exon 12 inclusion is marked below each lane with standard deviations (n = 3). A Western blot with anti-QKI-5 antibody indicates the expression of QKI-5 and QKI-5ΔKH proteins in cells cotransfected with pMAGPY and indicated amounts of QKI-5 expression constructs. The endogenous QKI-5 protein was indicated by the band in the pcDNA3 only lane.
Figure 3
Comparison of QKI-5 effects on alternative splicing in vivo. Splicing reporters: MAGPY (lanes 1–6), GABA γ2 (lanes 7–12), NMDA E21 (lanes 13–18); and protein expression plasmids, pcDNA QKI-5 and pcDNA PTB, were transfected into the nonneural cell line C2C12, and alternative splicing products were assayed by RT-PCR with primers (arrows) specific for sequences in the first and last exons. Exon-included and exon-skipped products are shown at left of each panel. Wedge indicates a 1:4 or 1:6 ratio of splicing reporter substrate to protein expression plasmid, left and right lanes, respectively. Control lanes (−) were transfected with empty pcDNA vector backbone. Structures of splicing reporter substrates are indicated schematically with exon and intron lengths in nucleotides (bottom right).
Figure 4
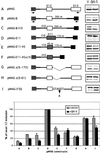
Sequence requirement for exon 12 inclusion and QKI-5 regulation. Mutations and deletions of MAG minigene are drawn schematically. MAG exons 12 and 13 are shown as white boxes and exon 11 as a cross-hatched box, and tat exons in the vector are shown in gray. Construct A, the wild-type pMAG. In constructs B–F, exon 12 or 11 and their flanking region were cloned between two tat exons. I10 refers to MAG intron 10. In constructs D–F, exon 12 is replaced with exon 11 (100 nt) or a modified exon 11 (45 nt). Constructs G and H represent two internal deletions downstream of exon 12. In construct I, the 5′ splice site of exon 12 was changed to the consensus GTAAGT. The arrows indicate primers. The right side represents the RT-PCR results of MAG minigene splicing products from COS7 cells cotransfected with these constructs and QKI-5 expression plasmid or the vector (V) alone. At the bottom is shown graphical comparison of the percentage of exon 12 inclusion from these mutations with SD (n = 3).
Figure 5
Sequence alignment between mouse and human MAG intron 12. The 170 nt immediately downstream of mouse MAG exon 12 is compared with the corespondent sequence in human MAG. The identical nucleotides are marked in gray boxes. The 53-nt QASE element is underlined.
Figure 6
Specific association of QKI-5 and QASE. (A) Diagrams of the RNA probe, E12QASE, ΔQASE, and competitor, EP160. (B) QKI-5 supershift assay. 32P-labeled E12QASE (native or heat-denatured) and ΔQASE (native) RNA probes were mixed with increasing amounts of HeLa nuclear extract (lanes 1–4, 9–11, 15–17). The amounts of anti-QKI-5 antibody or preimmune serum added to supershift reactions (lanes 5–8, 12, 13, 18–20) are indicated above. (C) Competition assay. Indicated amounts of HeLa nuclear extract were used in the gel mobility-shift assay. Used was 100-, 200-, or 400-fold of unlabeled RNA probe or competitor (lanes 5–10) in the competition reactions. (D) UV crosslinking assay of HeLa nuclear extract to native E12QASE probe (lanes 2–6). In lanes 3–6, 100- or 200-fold of competitors were added to the reactions before adding the probes. 35S-labeled in vitro translated QKI-5 protein was analyzed on the same gel (lane 1).
Similar articles
- Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein.
Zhao L, Mandler MD, Yi H, Feng Y. Zhao L, et al. Proc Natl Acad Sci U S A. 2010 Nov 2;107(44):19061-6. doi: 10.1073/pnas.1007487107. Epub 2010 Oct 18. Proc Natl Acad Sci U S A. 2010. PMID: 20956316 Free PMC article. - Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins.
Li Z, Zhang Y, Li D, Feng Y. Li Z, et al. J Neurosci. 2000 Jul 1;20(13):4944-53. doi: 10.1523/JNEUROSCI.20-13-04944.2000. J Neurosci. 2000. PMID: 10864952 Free PMC article. - The quakingviable mutation affects qkI mRNA expression specifically in myelin-producing cells of the nervous system.
Lu Z, Zhang Y, Ku L, Wang H, Ahmadian A, Feng Y. Lu Z, et al. Nucleic Acids Res. 2003 Aug 1;31(15):4616-24. doi: 10.1093/nar/gkg635. Nucleic Acids Res. 2003. PMID: 12888522 Free PMC article. - Molecular defects in the dysmyelinating mutant quaking.
Hardy RJ. Hardy RJ. J Neurosci Res. 1998 Feb 15;51(4):417-22. doi: 10.1002/(SICI)1097-4547(19980215)51:4<417::AID-JNR1>3.0.CO;2-F. J Neurosci Res. 1998. PMID: 9514195 Review. - QUAKING KH domain proteins as regulators of glial cell fate and myelination.
Larocque D, Richard S. Larocque D, et al. RNA Biol. 2005 Apr;2(2):37-40. doi: 10.4161/rna.2.2.1603. Epub 2005 Apr 9. RNA Biol. 2005. PMID: 17132940 Review.
Cited by
- Qki5 safeguards spinal motor neuron function by defining the motor neuron-specific transcriptome via pre-mRNA processing.
Hayakawa-Yano Y, Furukawa T, Matsuo T, Ogasawara T, Nogami M, Yokoyama K, Yugami M, Shinozaki M, Nakamoto C, Sakimura K, Koyama A, Ogi K, Onodera O, Takebayashi H, Okano H, Yano M. Hayakawa-Yano Y, et al. Proc Natl Acad Sci U S A. 2024 Sep 10;121(37):e2401531121. doi: 10.1073/pnas.2401531121. Epub 2024 Sep 3. Proc Natl Acad Sci U S A. 2024. PMID: 39226364 Free PMC article. - QKI-7 regulates expression of interferon-related genes in human astrocyte glioma cells.
Jiang L, Saetre P, Radomska KJ, Jazin E, Lindholm Carlström E. Jiang L, et al. PLoS One. 2010 Sep 29;5(9):e13079. doi: 10.1371/journal.pone.0013079. PLoS One. 2010. PMID: 20927331 Free PMC article. - Mechanisms Regulating Abnormal Circular RNA Biogenesis in Cancer.
Huang Y, Zhu Q. Huang Y, et al. Cancers (Basel). 2021 Aug 20;13(16):4185. doi: 10.3390/cancers13164185. Cancers (Basel). 2021. PMID: 34439339 Free PMC article. Review. - Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Teplova M, Hafner M, Teplov D, Essig K, Tuschl T, Patel DJ. Teplova M, et al. Genes Dev. 2013 Apr 15;27(8):928-40. doi: 10.1101/gad.216531.113. Genes Dev. 2013. PMID: 23630077 Free PMC article. - Sbp2l contributes to oligodendrocyte maturation through translational control in Tcf7l2 signaling.
Yugami M, Hayakawa-Yano Y, Ogasawara T, Yokoyama K, Furukawa T, Hara H, Hashikami K, Tsuji I, Takebayashi H, Araki S, Okano H, Yano M. Yugami M, et al. iScience. 2023 Nov 16;26(12):108451. doi: 10.1016/j.isci.2023.108451. eCollection 2023 Dec 15. iScience. 2023. PMID: 38213786 Free PMC article.
References
- Grabowski P J, Black D L. Prog Neurobiol. 2001;65:289–308. - PubMed
- Lopez A J. Annu Rev Genet. 1998;32:279–305. - PubMed
- Modrek B, Lee C. Nat Genet. 2002;30:13–19. - PubMed
- Sidman R L, Dickie M M, Appel S H. Science. 1964;144:309–311. - PubMed
- Cox R D, Hugill A, Shedlovsky A, Noveroske J K, Best S, Justice M J, Lehrach H, Dove W F. Genomics. 1999;57:333–341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials