Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability - PubMed (original) (raw)
Comparative Study
Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability
Anna V Roschke et al. Neoplasia. 2002 Jan-Feb.
Abstract
Most human tumors and tumor cell lines exhibit numerical and structural chromosomal abnormalities. The goal of this study was to determine the ongoing rates of structural and numerical instability in selected cancer cell lines and to investigate the consequences of these rates to karyotypic progression. We studied two colorectal (HCT-116 and HT-29) and two ovarian (SKOV-3 and OVCAR-8) cancer cell lines and their single cell subclones. We found that the signature karyotypes of all four cell lines were distinct and each aberrant. Whereas high rates of ongoing structural and/ or numerical chromosomal instability could be demonstrated in all cell lines, there was a relative stability of the consensus karyotype over many generations. No new clonal structural chromosomal reconfigurations emerged and the few numerical changes of karyotypes were restricted to abnormal chromosomes. This implies a kind of genomic optimization under the conditions of cell culture and suggests a link between genomic stabilization and cell propagation. We have been able to support this possibility by computer modeling. We did not observe a profound difference in the rates of numerical or structural instability in the cell lines with a replication error phenotype (RER+) versus the other cell lines.
Figures
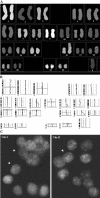
Figure 1
SKY, CGH, and FISH with centromere-specific probes of the cell line HCT-116, subclones A and B. (A) Karyotype of the HCT-116 cell line, subclone B, in classification colors. The only karyotypic difference between subclones A and B was the presence in the B of a balanced translocation involving chromosome 4 and der(18)t(17;18). (B) The average CGH ratio profiles for the near-diploid cell line HCT-116, subclone B. Note that all aberrations detected by SKY were also seen by CGH analysis, except for a balanced translocation between chromosomes 4 and der(18)t(17;18), and a loss of chromosome Y (normal female metaphase spreads were used for CGH experiments). CGH profiles for parental cell line HCT-116 and subclones A and B were identical. (C) Interphase FISH with chromosome-specific centromeric probes for chromosome 1 and 11 (HCT-116, subclone A). Arrowheads show nuclei with number of signals different from the modal number of chromosomes for that specific probe. Note that the modal number for chromosomes 1 and 11 was two for all three methods of analysis (SKY, CGH, and FISH).
Figure 2
SKY, CGH, and FISH with centromere-specific probes of cell line HT-29, subclone B. (A) Karyotype of the near-triploid HT-29 cell line, subclone B, in classification colors. The karyotype of this subclone was slightly different from the parental cell line. Three copies of chromosome 7 were present in the subclone, whereas the parental cell line had four copies. (B) The average CGH ratio profiles for the cell line HT-29, subclone B. Note that all aberrations detected by SKY were also seen by CGH analysis except a balanced translocation (6;14). (C) Interphase FISH with chromosome-specific centromeric probes for chromosomes 1, 11, and 17. Arrowheads show nuclei with number of signals different from the modal number of chromosomes for that specific probe. Note that the modal number for chromosomes 1 and 17 was three, and for chromosome 11 was four according to all three methods of analysis (SKY, CGH, and FISH). Also, the modal numbers for all other chromosomes studied in the HT-29, subclone B, were in complete agreement with FISH, SKY, and CGH.
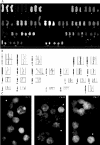
Figure 3
Spectral karyotype of the SKOV-3 parental cell line in display colors (A) and classification colors (B). Nonclonal chromosomal rearrangements (found in this cell only) are labeled with an asterisk (*).
Figure 4
(A) Spectral karyotype of the OVCAR-8 cell line in display colors. Chromosomes frequently undergoing additional rearrangements are indicated with arrows. (B) Examples of nonclonal variations of a clonal aberration. Nonclonal variants were collected from SKY-analyzed cells of the OVCAR-8 parental cell line and subclones.
Figure 5
The scheme for computer simulations.
Similar articles
- Possible causes of chromosome instability: comparison of chromosomal abnormalities in cancer cell lines with mutations in BRCA1, BRCA2, CHK2 and BUB1.
Grigorova M, Staines JM, Ozdag H, Caldas C, Edwards PA. Grigorova M, et al. Cytogenet Genome Res. 2004;104(1-4):333-40. doi: 10.1159/000077512. Cytogenet Genome Res. 2004. PMID: 15162061 - Distinct patterns of structural and numerical chromosomal instability characterize sporadic ovarian cancer.
Bayani J, Paderova J, Murphy J, Rosen B, Zielenska M, Squire JA. Bayani J, et al. Neoplasia. 2008 Oct;10(10):1057-65. doi: 10.1593/neo.08584. Neoplasia. 2008. PMID: 18813350 Free PMC article. - Comprehensive measurement of chromosomal instability in cancer cells: combination of fluorescence in situ hybridization and cytokinesis-block micronucleus assay.
Camps J, Ponsa I, Ribas M, Prat E, Egozcue J, Peinado MA, Miró R. Camps J, et al. FASEB J. 2005 May;19(7):828-30. doi: 10.1096/fj.04-2276fje. Epub 2005 Mar 9. FASEB J. 2005. PMID: 15760839 - Molecular cytogenetic characterization of pancreas cancer cell lines reveals high complexity chromosomal alterations.
Griffin CA, Morsberger L, Hawkins AL, Haddadin M, Patel A, Ried T, Schrock E, Perlman EJ, Jaffee E. Griffin CA, et al. Cytogenet Genome Res. 2007;118(2-4):148-56. doi: 10.1159/000108295. Cytogenet Genome Res. 2007. PMID: 18000365 Review. - Chromosomal imbalances in oral squamous cell carcinoma: examination of 31 cell lines and review of the literature.
Martin CL, Reshmi SC, Ried T, Gottberg W, Wilson JW, Reddy JK, Khanna P, Johnson JT, Myers EN, Gollin SM. Martin CL, et al. Oral Oncol. 2008 Apr;44(4):369-82. doi: 10.1016/j.oraloncology.2007.05.003. Epub 2007 Aug 2. Oral Oncol. 2008. PMID: 17681875 Free PMC article. Review.
Cited by
- Imaging Unique DNA Sequences in Individual Cells Using a CRISPR-Cas9-Based, Split Luciferase Biosensor.
Heath NG, O'Geen H, Halmai NB, Corn JE, Segal DJ. Heath NG, et al. Front Genome Ed. 2022 Mar 25;4:867390. doi: 10.3389/fgeed.2022.867390. eCollection 2022. Front Genome Ed. 2022. PMID: 35403097 Free PMC article. - Reverting to single-cell biology: The predictions of the atavism theory of cancer.
Bussey KJ, Davies PCW. Bussey KJ, et al. Prog Biophys Mol Biol. 2021 Oct;165:49-55. doi: 10.1016/j.pbiomolbio.2021.08.002. Epub 2021 Aug 8. Prog Biophys Mol Biol. 2021. PMID: 34371024 Free PMC article. - Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells.
Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Upender MB, et al. Cancer Res. 2004 Oct 1;64(19):6941-9. doi: 10.1158/0008-5472.CAN-04-0474. Cancer Res. 2004. PMID: 15466185 Free PMC article. - miR-214-mediated downregulation of RNF8 induces chromosomal instability in ovarian cancer cells.
Wang Z, Yin H, Zhang Y, Feng Y, Yan Z, Jiang X, Bukhari I, Iqbal F, Cooke HJ, Shi Q. Wang Z, et al. Cell Cycle. 2014;13(22):3519-28. doi: 10.4161/15384101.2014.958413. Cell Cycle. 2014. PMID: 25483088 Free PMC article. - Genomic analysis of genetic heterogeneity and evolution in high-grade serous ovarian carcinoma.
Cooke SL, Ng CK, Melnyk N, Garcia MJ, Hardcastle T, Temple J, Langdon S, Huntsman D, Brenton JD. Cooke SL, et al. Oncogene. 2010 Sep 2;29(35):4905-13. doi: 10.1038/onc.2010.245. Epub 2010 Jun 28. Oncogene. 2010. PMID: 20581869 Free PMC article.
References
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. - PubMed
- Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. - PubMed
- Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. - PubMed
- Ilyas M, Straub J, Tomlinson IP, Bodmer WF. Genetic pathways in colorectal and other cancers. Eur J Cancer. 1999;35:1986–2002. - PubMed
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical