Applying a highly specific and reproducible cDNA RDA method to clone garlic up-regulated genes in human gastric cancer cells - PubMed (original) (raw)
Applying a highly specific and reproducible cDNA RDA method to clone garlic up-regulated genes in human gastric cancer cells
Yong Li et al. World J Gastroenterol. 2002 Apr.
Abstract
Aim: To develop and optimize cDNA representational difference analysis (cDNA RDA) method and to identify and clone garlic up-regulated genes in human gastric cancer (HGC) cells.
Methods: We performed cDNA RDA method by using abundant double-stranded cDNA messages provided by two self-constructed cDNA libraries (Allitridi-treated and paternal HGC cell line BGC823 cells cDNA libraries respectively). Bam H I and Xho I restriction sites harbored in the library vector were used to select representations. Northern and Slot blots analyses were employed to identify the obtained difference products.
Results: Fragments released from the cDNA library vector after restriction endonuclease digestion acted as good marker indicating the appropriate digestion degree for library DNA. Two novel expressed sequence tags (ESTs) and a recombinant gene were obtained. Slot blots result showed a 8-fold increase of glia-derived nexin/protease nexin 1 (GDN/PN1) gene expression level and 4-fold increase of hepatitis B virus x-interacting protein (XIP) mRNA level in BGC823 cells after Allitridi treatment for 72h.
Conclusion: Elevated levels of GDN/PN1 and XIP mRNAs induced by Allitridi provide valuable molecular evidence for elucidating the garlic's efficacies against neurodegenerative and inflammatory diseases. Isolation of a recombinant gene and two novel ESTs further show cDNA RDA based on cDNA libraries to be a powerful method with high specificity and reproducibility in cloning differentially expressed genes.
Figures
Figure 1
Schematic diagram of preparing representations for cDNA RDA based on cDNA libraries. Using _Bam_H I alone (A) or _Bam_H I and Xho I together (B) to digest library DNAs. Black boxes represent insert cDNAs of cDNA libraries. White boxes represent two arms of cDNA library vector (λZAP II vector). “B” epresents _Bam_H I and “X” Xho I.
Figure 2
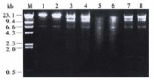
Agarose gel electrophoresis of digestion products of Alli823 (lanes 1, 3, 5) and BGC823 (lanes 2, 4, 6) cDNA library DNAs digested with _Bam_H I at 37 °C for 2 (lanes 1, 2), 4 (lanes 3, 4), and 12 h (lanes 5, 6) respectively. The digestion products of Alli823 (lane 7) and BGC823 (lane 8) library DNAs digested with _Bam_H I and Xho I together (A fragment, 850 bp in size, appeared in the digestion products). λPhage/_Hin_d III size marker (lane M).
Figure 3
Agarose gel electrophoresis of amplicons derived from different enzyme digestions. A: The amplicons obtained by using _Bam_H I to digest Alli823 (lane 1) and BGC823 (lane 2) cDNA library DNAs respectively; B: The amplicons obtained by using _Bam_H I and Xho I together to digest Alli823 (lane 1) and BGC823 (lane 2) library DNAs respectively. 1 kb size marker (lane M).
Figure 4
A: Agarose gel electrophoresis of reamplified difference products SH1-4 (lanes 1-4 respectively). PCR product of GAPDH (400 bp in size) used as quantity control (lane G); B: Slot blots analysis showing differentially expressed cDNAs. Reverse transcription products (first stranded cDNA) of total RNA of BGC823 and Alli823 cells used as probe respectively; C: The degree to which each cDNA was up-regulated.
Figure 5
Sequences of two novel ESTs (SH1 and SH4) isolated by cDNA RDA based on cDNA libraries.
Figure 6
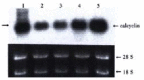
Northern blotting result showing the expression level of calcyclin gene in human gastric cancer cell lines BGC823, MGC803, PAMC82, SGC7901 and MKN45 (lanes 1-5) respectively.The arrow showing the novel transcript derived from the recombinant gene merged from calcyclin gene and HNRPA0 gene.
Similar articles
- Construction of cDNA representational difference analysis based on two cDNA libraries and identification of garlic inducible expression genes in human gastric cancer cells.
Li Y, Yang L, Cui JT, Li WM, Guo RF, Lu YY. Li Y, et al. World J Gastroenterol. 2002 Apr;8(2):208-12. doi: 10.3748/wjg.v8.i2.208. World J Gastroenterol. 2002. PMID: 11925593 Free PMC article. - Effects of allitridi on cell cycle arrest of human gastric cancer cells.
Ha MW, Ma R, Shun LP, Gong YH, Yuan Y. Ha MW, et al. World J Gastroenterol. 2005 Sep 21;11(35):5433-7. doi: 10.3748/wjg.v11.i35.5433. World J Gastroenterol. 2005. PMID: 16222732 Free PMC article. - [Effect of allitridi on cyclin D1 and p27(Kip1) protein expression in gastric carcinoma BGC823 cells].
Lan H, Lu YY. Lan H, et al. Ai Zheng. 2003 Dec;22(12):1268-71. Ai Zheng. 2003. PMID: 14693049 Chinese. - Anticancer effects of diallyl trisulfide derived from garlic.
Seki T, Hosono T, Hosono-Fukao T, Inada K, Tanaka R, Ogihara J, Ariga T. Seki T, et al. Asia Pac J Clin Nutr. 2008;17 Suppl 1:249-52. Asia Pac J Clin Nutr. 2008. PMID: 18296348 Review.
Cited by
- Effects of allicin on both telomerase activity and apoptosis in gastric cancer SGC-7901 cells.
Sun L, Wang X. Sun L, et al. World J Gastroenterol. 2003 Sep;9(9):1930-4. doi: 10.3748/wjg.v9.i9.1930. World J Gastroenterol. 2003. PMID: 12970878 Free PMC article. - Cloning of human 15ku selenoprotein gene from H9 T cells.
Nan KJ, Li CL, Wei YC, Sui CG, Jing Z, Qin HX, Zhao LJ, Pan BR. Nan KJ, et al. World J Gastroenterol. 2003 Aug;9(8):1777-80. doi: 10.3748/wjg.v9.i8.1777. World J Gastroenterol. 2003. PMID: 12918119 Free PMC article. - Comparative proteomic analysis of proteins involved in the tumorigenic process of seminal vesicle carcinoma in transgenic mice.
Chang WC, Chou CK, Tsou CC, Li SH, Chen CH, Zhuo YX, Hsu WL, Chen CH. Chang WC, et al. Int J Proteomics. 2010;2010:726968. doi: 10.1155/2010/726968. Epub 2010 May 12. Int J Proteomics. 2010. PMID: 22084680 Free PMC article. - Effect of apoptosis on gastric adenocarcinoma cell line SGC-7901 induced by cis-9, trans-11-conjugated linoleic acid.
Liu JR, Chen BQ, Yang YM, Wang XL, Xue YB, Zheng YM, Liu RH. Liu JR, et al. World J Gastroenterol. 2002 Dec;8(6):999-1004. doi: 10.3748/wjg.v8.i6.999. World J Gastroenterol. 2002. PMID: 12439913 Free PMC article. - Inhibition of beta-ionone on SGC-7901 cell proliferation and upregulation of metalloproteinases-1 and -2 expression.
Liu JR, Yang BF, Chen BQ, Yang YM, Dong HW, Song YQ. Liu JR, et al. World J Gastroenterol. 2004 Jan 15;10(2):167-71. doi: 10.3748/wjg.v10.i2.167. World J Gastroenterol. 2004. PMID: 14716815 Free PMC article.
References
- Yoon DY, Buchler P, Saarikoski ST, Hines OJ, Reber HA, Hankinson O. Identification of genes differentially induced by hypoxia in pancreatic cancer cells. Biochem Biophys Res Commun. 2001;288:882–886. - PubMed
- Graveel CR, Jatkoe T, Madore SJ, Holt AL, Farnham PJ. Expression profiling and identification of novel genes in hepatocellular carcinomas. Oncogene. 2001;20:2704–2712. - PubMed
- Seidita G, Polizzi D, Costanzo G, Costa S, Di Leonardo A. Differential gene expression in p53-mediated G (1) arrest of human fibroblasts after gamma-irradiation or N-phosphoacetyl-L-aspartate treatment. Carcinogenesis. 2000;21:2203–2210. - PubMed
- Xu W, Wang S, Wang G, Wei H, He F, Yang X. Identification and characterization of differentially expressed genes in the early response phase during liver regeneration. Biochem Biophys Res Commun. 2000;278:318–325. - PubMed
- Davenport J, Neale GA, Goorha R. Identification of genes potentially involved in LMO2-induced leukemogenesis. Leukemia. 2000;14:1986–1996. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous