Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication - PubMed (original) (raw)
Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication
Volker M Stucke et al. EMBO J. 2002.
Abstract
Budding yeast Mps1p kinase has been implicated in both the duplication of microtubule-organizing centers and the spindle assembly checkpoint. Here we show that hMps1, the human homolog of yeast Mps1p, is a cell cycle-regulated kinase with maximal activity during M phase. hMps1 localizes to kinetochores and its activity and phosphorylation state increase upon activation of the mitotic checkpoint. By antibody microinjection and siRNA, we demonstrate that hMps1 is required for human cells to undergo checkpoint arrest in response to microtubule depolymerization. In contrast, centrosome (re-)duplication as well as cell division occur in the absence of hMps1. We conclude that hMps1 is required for the spindle assembly checkpoint but not for centrosome duplication.
Figures
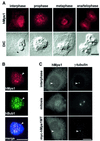
Fig. 1. Cell cycle regulation of hMps1 protein and activity levels. (A) Characterization of hMps1 antibodies. Cell extracts from exponentially growing HeLa cells were separated by SDS–PAGE and probed by western blotting with polyclonal, affinity-purified anti-hMps1 antibody (poly), and with three different anti-hMps1 mAbs (N1, N2 and C1). A non-immune IgG1 was used for control. The migration of hMps1 is shown on the left and molecuar weight markers are indicated on the right. The schematic representation (top) shows the distribution of epitopes recognized by the different mAbs on hMps1, with the catalytic domain marked in gray. (B–D) HeLa cells were released from a thymidine/aphidicolin double block in early S phase, samples were taken for FACS (B), western blot analysis (C) and kinase activity measurements (D) at the time points indicated. For comparison, asynchronously growing (asn) and nocodazole-arrested (noc) cells were analyzed in parallel. (E) hMps1 is phosphorylated specifically upon checkpoint engagement. Cell extracts were prepared from cells labeled in vivo with [32P]inorganic phosphate, and equal amounts of extracts were used for immunoprecipitations with hMps1 antibody or non-immune IgG1. Following SDS–PAGE, recovery of hMps1 protein was monitored by western blotting using mAb N1 (upper panel) and incorporation of 32P into hMps1 visualized by autoradiography (lower panel).
Fig. 2. Endogenous hMps1 transiently associates with kinetochores in U2OS cells. (A) Exponentially growing U2OS cells were fixed and permeabilized simultaneously with formaldehyde/Triton X-100 and analyzed by indirect immunofluorescence microscopy using anti-hMps1 mAb N1. Corresponding differential interference contrast (DIC) micrographs are shown in the lower panels. Bars = 10 µm. (B) Kinetochores were identified by double staining with anti-hMps1 mAb N1 and rabbit anti-hBub1 antibodies (arrowheads) after fixation as described in (A). DNA was counterstained using DAPI. In the merged panel, anti-hMps1 staining is in red and anti-hBub1 staining in green. Bar = 10 µm. (C) Neither endogenous hMps1 nor ectopically expressed hMps1 localize to centrosomes (arrowheads) in U2OS cells. Interphasic (upper panel) and mitotic (middle panel) U2OS cells were fixed with methanol and analyzed with antibodies against hMps1 (mAb N1; left) and γ-tubulin (right). The bottom panel shows the localization of wild-type, myc-tagged hMps1, after transient expression in U2OS cells and visualization with anti-myc (9E10) antibody (left), again in direct comparison with γ-tubulin localization (right). Bar = 10 µm.
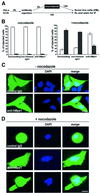
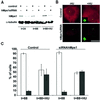
Fig. 3. hMps1 is an essential component of the spindle assembly checkpoint. (A) A schematic illustration of the experimental protocol used for microinjection experiments. (B) Histograms comparing the morphologies of HeLa cells after cytoplasmic injection of non-immune IgG1 or anti-hMps1 antibodies and subsequent incubation in the absence (left panel) or presence of nocodazole (right panel). Open bars indicate the percentage of injected cells with a flattened, interphasic morphology, whereas black bars indicate the proportion of cells with a rounded morphology (i.e. mitotically arrested cells). Approximately 120–150 injected cells were counted for each experiment. Shown are the averages of three independent experiments, with standard deviations. (C and D) Single, widely spaced HeLa cells were injected with control IgG1 or anti-hMps1 antibodies (as indicated), followed by either a 14 h incubation in the absence of nocodazole (C), or a 12 h incubation with nocodazole (D), before fixation with paraformaldehyde solution. Injected cells were visualized using an anti-mouse IgG secondary antibody and DNA was stained by DAPI. Representative examples of injected HeLa cells are shown. Note that the presence of daughter cells indicates successful cell division. Bars = 10 µm.
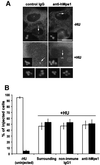
Fig. 4. Microinjection of anti-hMps1 antibodies does not block centrosome duplication. (A) U2OS cells were pre-synchronized for 16 h in 15 mM hydroxyurea (HU) and injected into the cytoplasm with either control IgG1 (left panels) or anti-hMps1 antibodies (right panels). Injections were scattered (i.e. only one cell per microscopic field) for accurate counting. Cells were then incubated for 24 h in the absence of HU (upper panels) or for 64 h in the presence of 15 mM HU (lower panels), before they were methanol fixed and double-stained using an anti-mouse IgG secondary antibody to visualize injected antibodies, and anti-γ-tubulin to visualize centrosomes. Note that cells had divided properly in the absence of HU (upper panels), whereas centrosome reduplication had occurred in the presence of HU-induced S phase arrest, regardless of anti-hMps1 antibody injection. Bar = 10 µm. (B) Quantitative analysis of centrosome reduplication. The histogram indicates the percentages of cells with normal numbers (1–2) of centrosomes (open bars) or extra copies (>2) of centrosomes (black bars). Incubation of U2OS cells with or without HU, as well as the identification of injected cells and centrosome staining, were performed as described in (A). Approximately 80–120 injected cells were counted for each experiment. Shown are the averages of three independent experiments ± SD.
Fig. 5. Silencing of hMps1 by siRNA duplex abolishes the spindle assembly checkpoint. (A) Western blotting shows effective silencing of hMps1 in HeLa S3 cells (transfection efficiency = 90%). Following mock transfection or transfection with hMps1 siRNA duplex, cell extracts were prepared at the time points indicated. Equal amounts of protein were separated by SDS–PAGE and probed by western blotting with anti-hMps1 mAb N1 (upper panel) and anti-α-tubulin antibody as a loading control (lower panel). In the sample marked noc, nocodazole (50 ng/ml) was added at t = 48 h for an additional 24 h. (B) Immunofluorescence microscopy shows effective silencing of hMps1 in HeLaS3 cells. Cells were transfected as described in (A), fixed and permeabilized with paraformaldehyde/Triton X-100, and stained for hMps1 (upper panels) and DNA (lower panels) using anti-hMps1 mAb N1 and DAPI, respectively. Bar = 10 µm. (C) Analysis of HeLa S3 cells after mock transfection (left) or hMps1 silencing by siRNA duplex (right) and subsequent exposure to spindle damage. Nocodazole (50 ng/ml) was added 48 h post-transfection for 24 h before cells were analyzed. Upper panels: phase-contrast pictures. Lower panels: cells stained for hMps1 (left) and DNA (right). Bars = 1 µm (upper panels) and 10 µm (lower panels). (D) Cell extracts, prepared from the samples described in (C), were probed by western blotting with anti-cyclin B1 and anti-p27Kip1 antibodies, respectively. Anti-α-tubulin was used as a loading control.
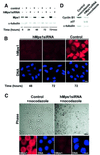
Fig. 6. Silencing of hMps1 by siRNA duplex does not interfere with centrosome (re-)duplication. (A) Western blotting demonstrates effective silencing of hMps1 in U2OS cells (transfection efficiency = 70%). Cells were transfected with GL-2 control or hMps1 siRNA duplexes, and extracts were prepared at the times indicated. Equal amounts of protein were resolved by SDS–PAGE and probed by western blotting with anti-hMps1 mAb N1 (upper panel) and anti-α-tubulin as a loading control (lower panel). In the sample marked t = 88 h + HU, hydroxyurea (15 mM) was added at t = 24 h for an additional 64 h. (B) Immunofluorescence microscopy shows effective silencing of hMps1 in U2OS cells. Cells were transfected as described in (A), fixed with methanol and stained for hMps1 (red) and γ-tubulin (green). Bar = 10 µm. (C) Histogram comparing the efficiency of centrosome reduplication in the samples described above. Open bars indicate the percentage of cells with normal numbers (1–2) of centrosomes, whereas black bars denote cells with multiple centrosomes (≥3). Only cells displaying low levels of hMps1 staining were counted. Shown are the averages of three independent experiments (counting ∼200 cells each) ± SD.
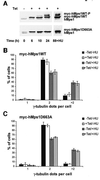
Fig. 7. Overexpression of wild-type myc-hMps1 (myc-hMps1WT) and myc-hMps1D663A does not affect centrosome reduplication. (A) Tetracycline-inducible expression of myc-hMps1WT and myc-hMps1D663A. Stably transfected U2OS-TRex cells were induced with tetracycline (1 µg/ml), and levels of hMps1 determined by immunoblotting. (B) Myc-hMps1WT does not enhance centrosome (re-)duplication. After an initial 24 h induction of myc-hMps1WT by tetracycline (1 µg/ml) in the absence of HU, myc-hMps1WT expression was continued for 64 h in the presence or absence of 15 mM HU, as indicated. Cells were then fixed with methanol and stained for myc-hMps1 and γ-tubulin, and the numbers of centrosomes counted. The histogram shows the results of three independent experiments ± SD (at least 200 cells were counted in each experiment). (C) Myc-hMps1D663A does not block centrosome (re-)duplication. Induction of myc-hMps1D663A and analysis of cells were performed as described in (B).
Similar articles
- Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression.
Fisk HA, Mattison CP, Winey M. Fisk HA, et al. Proc Natl Acad Sci U S A. 2003 Dec 9;100(25):14875-80. doi: 10.1073/pnas.2434156100. Epub 2003 Dec 1. Proc Natl Acad Sci U S A. 2003. PMID: 14657364 Free PMC article. - Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores.
Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Liu ST, et al. Mol Biol Cell. 2003 Apr;14(4):1638-51. doi: 10.1091/mbc.02-05-0074. Mol Biol Cell. 2003. PMID: 12686615 Free PMC article. - Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2.
Martin-Lluesma S, Stucke VM, Nigg EA. Martin-Lluesma S, et al. Science. 2002 Sep 27;297(5590):2267-70. doi: 10.1126/science.1075596. Science. 2002. PMID: 12351790 - Centrosome duplication: three kinases come up a winner!
Hinchcliffe EH, Sluder G. Hinchcliffe EH, et al. Curr Biol. 2001 Sep 4;11(17):R698-701. doi: 10.1016/s0960-9822(01)00412-2. Curr Biol. 2001. PMID: 11553343 Review. - The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle.
Wang G, Jiang Q, Zhang C. Wang G, et al. J Cell Sci. 2014 Oct 1;127(Pt 19):4111-22. doi: 10.1242/jcs.151753. Epub 2014 Aug 15. J Cell Sci. 2014. PMID: 25128564 Review.
Cited by
- Ab ovo or de novo? Mechanisms of centriole duplication.
Loncarek J, Khodjakov A. Loncarek J, et al. Mol Cells. 2009 Feb 28;27(2):135-42. doi: 10.1007/s10059-009-0017-z. Epub 2009 Feb 20. Mol Cells. 2009. PMID: 19277494 Free PMC article. Review. - Haploid genetic screens identify genetic vulnerabilities to microtubule-targeting agents.
Gerhards NM, Blomen VA, Mutlu M, Nieuwenhuis J, Howald D, Guyader C, Jonkers J, Brummelkamp TR, Rottenberg S. Gerhards NM, et al. Mol Oncol. 2018 Jun;12(6):953-971. doi: 10.1002/1878-0261.12307. Epub 2018 May 1. Mol Oncol. 2018. PMID: 29689640 Free PMC article. - Phosphorylation at threonine 288 by cell cycle checkpoint kinase 2 (CHK2) controls human monopolar spindle 1 (Mps1) kinetochore localization.
Yeh CW, Yu ZC, Chen PH, Cheng YC, Shieh SY. Yeh CW, et al. J Biol Chem. 2014 May 30;289(22):15319-27. doi: 10.1074/jbc.M114.552273. Epub 2014 Apr 24. J Biol Chem. 2014. PMID: 24764296 Free PMC article. - Centrosomes and cancer: revisiting a long-standing relationship.
Gönczy P. Gönczy P. Nat Rev Cancer. 2015 Nov;15(11):639-52. doi: 10.1038/nrc3995. Nat Rev Cancer. 2015. PMID: 26493645 Review. - A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B.
Nijenhuis W, von Castelmur E, Littler D, De Marco V, Tromer E, Vleugel M, van Osch MH, Snel B, Perrakis A, Kops GJ. Nijenhuis W, et al. J Cell Biol. 2013 Apr 15;201(2):217-31. doi: 10.1083/jcb.201210033. Epub 2013 Apr 8. J Cell Biol. 2013. PMID: 23569217 Free PMC article.
References
- Abrieu A., Magnaghi-Jaulin,L., Kahana,J.A., Peter,M., Castro,A., Vigneron,S., Lorca,T., Cleveland,D.W. and Labbe,J.C. (2001) Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell, 106, 83–93. - PubMed
- Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. - PubMed
- Blangy A., Lane,H.A., d’Herin,P., Harper,M., Kress,M. and Nigg,E.A. (1995) Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell, 83, 1159–1169. - PubMed
- Clute P. and Pines,J. (1999) Temporal and spatial control of cyclin B1 destruction in metaphase. Nature Cell Biol., 1, 82–87. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases