The Paf1 complex physically and functionally associates with transcription elongation factors in vivo - PubMed (original) (raw)
The Paf1 complex physically and functionally associates with transcription elongation factors in vivo
Sharon L Squazzo et al. EMBO J. 2002.
Abstract
We are using biochemical and genetic approaches to study Rtf1 and the Spt4-Spt5 complex, which independently have been implicated in transcription elongation by RNA polymerase II. Here, we report a remarkable convergence of these studies. First, we purified Rtf1 and its associated yeast proteins. Combining this approach with genetic analysis, we show that Rtf1 and Leo1, a protein of unknown function, are members of the RNA polymerase II-associated Paf1 complex. Further analysis revealed allele-specific genetic interactions between Paf1 complex members, Spt4-Spt5, and Spt16-Pob3, the yeast counterpart of the human elongation factor FACT. In addition, we independently isolated paf1 and leo1 mutations in an unbiased genetic screen for suppressors of a cold-sensitive spt5 mutation. These genetic interactions are supported by physical interactions between the Paf1 complex, Spt4-Spt5 and Spt16-Pob3. Finally, we found that defects in the Paf1 complex cause sensitivity to 6-azauracil and diminished PUR5 induction, properties frequently associated with impaired transcription elongation. Taken together, these data suggest that the Paf1 complex functions during the elongation phase of transcription in conjunction with Spt4-Spt5 and Spt16-Pob3.
Figures
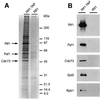
Fig. 1. Rtf1–TAP co-purifies with Paf1, Cdc73, Ctr9, Leo1, Spt5 and Rpb1. Yeast strains expressing untagged and TAP-tagged forms of Rtf1 were processed in parallel as described in Materials and methods. (A) Silver-stained gel of purified proteins derived from ∼150 mg of protein extract. Identification of bands as Rtf1, Paf1 and Cdc73 was based upon immunoblot analyses carried out in parallel. Similar assignments were not made for Ctr9 and Leo1 (predicted masses of ∼125 and 54 kDa, respectively) because antibodies to these proteins were not available and because individual bands were not excised for our mass spectrometry analysis. (B) Immunoblots of purified proteins, visualized with antisera directed against Rtf1, Paf1, Cdc73 and Spt5. The monoclonal antibody 8WG16 was used to probe for Rpb1.
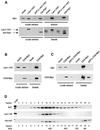
Fig. 2. Rtf1 and Leo1 are members of the Paf1 complex. (A) Rtf1 co-immunoprecipitates with Leo1-HA1 and HA1-Paf1. Anti-HA1 immunoprecipitations were performed from extracts of strains lacking an HA1-tagged protein (mock, GHY1102), containing either HA1-tagged Paf1 (GHY1147) or Leo1 (GHY1141), and from a LEO1-HA1 paf1Δ strain (GHY1146). Proteins that precipitated with the HA1 antibody-bound beads were eluted with 1 M KCl (elution) and separated by SDS–PAGE. Proteins that remained bound to the beads after the 1 M KCl elution were liberated by boiling in sample buffer (beads). The HA1-tagged proteins and Rtf1 were visualized by immunoblotting. (B) Leo1-HA1 co-immunoprecipitates with Ctr9-Myc. Immunoprecipitations were performed on extracts of a Leo1-HA1 (mock, GHY1092) and a Ctr9-Myc Leo1-HA1 strain (GHY1184). (C) Rtf1 co- immunoprecipitates with Ctr9-Myc. Immunoprecipitations were performed on extracts of a strain lacking a myc tag (mock, GHY1002), a Ctr9-Myc strain (GHY1184) and a paf1Δ Ctr9-Myc strain (GHY1222). (D) Paf1, Leo1 and Rtf1 co-fractionate on a gel filtration column. Whole-cell extracts derived from a Leo1-HA1 strain (GHY1140) and a HA1-Paf1 strain (GHY1147) were fractionated on a Superose 6 column, and the indicated fractions from the eluates were analyzed by immunoblotting. Each experiment was performed at least twice, with peak elution of each protein reproducibly occurring in fraction 10.
Fig. 3. Mutant phenotypes shared by Paf1 complex members. (A) Strains with the indicated genotypes and containing the his4-912δ and lys2-128δ insertions were grown on YPD media, replica-plated to minimal media supplemented with histidine, lysine, leucine, uracil and tryptophan (complete), and SC-his and SC-lys plates, and incubated for 3 days at 30°C. Strains: WT, FY118; rtf1Δ, KY522; paf1Δ, GHY917; cdc73Δ, GHY1083; leo1Δ, GHY240; ctr9Δ, GHY1157. (B) Strains with the indicated genotypes were grown on YPD media and then replica-plated to SC-ura media lacking or containing 50 µg/ml 6AU. Photographs were taken after 2 and 4 days of growth at 30°C. Strains: wild-type, KY286; rtf1Δ, KY425; paf1Δ, KY686; cdc73Δ, KY689; leo1Δ, KY690; ctr9Δ, KY695.
Fig. 3. Mutant phenotypes shared by Paf1 complex members. (A) Strains with the indicated genotypes and containing the his4-912δ and lys2-128δ insertions were grown on YPD media, replica-plated to minimal media supplemented with histidine, lysine, leucine, uracil and tryptophan (complete), and SC-his and SC-lys plates, and incubated for 3 days at 30°C. Strains: WT, FY118; rtf1Δ, KY522; paf1Δ, GHY917; cdc73Δ, GHY1083; leo1Δ, GHY240; ctr9Δ, GHY1157. (B) Strains with the indicated genotypes were grown on YPD media and then replica-plated to SC-ura media lacking or containing 50 µg/ml 6AU. Photographs were taken after 2 and 4 days of growth at 30°C. Strains: wild-type, KY286; rtf1Δ, KY425; paf1Δ, KY686; cdc73Δ, KY689; leo1Δ, KY690; ctr9Δ, KY695.
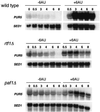
Fig. 4. rtf1Δ and paf1Δ strains are defective in PUR5 induction. Northern analysis of PUR5 transcription was performed on total RNA samples prepared from wild-type (KY589), rtf1Δ (KY453) and paf1Δ (KY687) cells. The strains were grown to ∼1 × 107 cells/ml in SC-ura media and then divided. 6AU was added to a final concentration of 75 µg/ml to one half of the culture and both cultures were grown at 30°C for the indicated times, in hours, prior to RNA isolation. Each lane contained 10 µg of total RNA. The same filters that were probed for PUR5 mRNA were stripped and probed for SED1 mRNA to serve as a loading control.
Fig. 5. Suppression of the spt5Cs– growth defect by paf1 and leo1 mutations. Strains with the indicated genotypes were replica-plated to YPD media and incubated for 2 (30°C) or 4 (15°C) days. Strains: _spt5Cs_–, GHY92; wild type, FY653; _paf1-49 spt5Cs_–, GHY144; _leo1-43 spt5Cs_–, GHY141.
Fig. 6. Transcription elongation factors associate with the Paf1 complex. (A) Spt5, Spt16 and Pob3 co-immunoprecipitate with HA1-Paf1 and Leo1-HA1. Anti-HA1 immunoprecipitations were performed from extracts of a strain lacking an HA1 epitope tag (mock, GHY1102), strains containing Leo1-HA1 (GHY1184), HA1-Paf1 (GHY1147) and from a paf1Δ Leo1-HA1 strain (GHY1146). The immunoprecipitates were analyzed by immunoblotting for Spt5, Spt16 and Pob3 as indicated. (B) Spt5 co-immunoprecipitates with Ctr9. Anti-Myc immunoprecipitations were performed on extracts of a strain lacking a Myc tag (mock, GHY1002), a Ctr9-Myc strain (GHY1184) and a paf1Δ Ctr9-Myc strain (GHY1222). Bound proteins were eluted from the anti-Myc beads by a 1 M salt elution (elution) and then by boiling the beads in sample buffer (beads). The immunoprecipitates were analyzed by immunoblotting for Spt5 and Ctr9, with an anti-Myc antibody, as indicated. (C) Whole-cell extracts were prepared from yeast strain FY1639 and used for immunoprecipitations of Rtf1. Western blots were probed with anti-Spt5, anti-Rtf1 and anti-TBP antisera. A 20 µg aliquot of whole-cell extract, 7 µg of unbound protein and three-fifths of the immunoprecipitated sample (beads) were analyzed by immunoblotting with the indicated antibodies. (D) Growth defects in paf1Δ dst1Δ and paf1Δ spt16-197 double mutants. Serial dilutions of cells with the indicated genotypes were spotted onto YPD media and grown for 2 days at 30°C. Strains: wild-type, FY653; paf1Δ, GHY917; dst1Δ, GHY285; spt16-197, GHY1059; paf1Δ dst1Δ, GHY1009; paf1Δ spt16-197, GHY1228.
Similar articles
- RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach.
Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. Krogan NJ, et al. Mol Cell Biol. 2002 Oct;22(20):6979-92. doi: 10.1128/MCB.22.20.6979-6992.2002. Mol Cell Biol. 2002. PMID: 12242279 Free PMC article. - Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae.
Hartzog GA, Wada T, Handa H, Winston F. Hartzog GA, et al. Genes Dev. 1998 Feb 1;12(3):357-69. doi: 10.1101/gad.12.3.357. Genes Dev. 1998. PMID: 9450930 Free PMC article. - Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes.
Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Simic R, et al. EMBO J. 2003 Apr 15;22(8):1846-56. doi: 10.1093/emboj/cdg179. EMBO J. 2003. PMID: 12682017 Free PMC article. - Transcription elongation: the 'Foggy' is liftingellipsis.
Zorio DA, Bentley DL. Zorio DA, et al. Curr Biol. 2001 Feb 20;11(4):R144-6. doi: 10.1016/s0960-9822(01)00063-x. Curr Biol. 2001. PMID: 11250170 Review. - The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation.
Hartzog GA, Fu J. Hartzog GA, et al. Biochim Biophys Acta. 2013 Jan;1829(1):105-15. doi: 10.1016/j.bbagrm.2012.08.007. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982195 Free PMC article. Review.
Cited by
- Mechanisms and Functions of the RNA Polymerase II General Transcription Machinery during the Transcription Cycle.
Archuleta SR, Goodrich JA, Kugel JF. Archuleta SR, et al. Biomolecules. 2024 Feb 1;14(2):176. doi: 10.3390/biom14020176. Biomolecules. 2024. PMID: 38397413 Free PMC article. Review. - Identification of Mutant Versions of the Spt16 Histone Chaperone That Are Defective for Transcription-Coupled Nucleosome Occupancy in Saccharomyces cerevisiae.
Hainer SJ, Charsar BA, Cohen SB, Martens JA. Hainer SJ, et al. G3 (Bethesda). 2012 May;2(5):555-67. doi: 10.1534/g3.112.002451. Epub 2012 May 1. G3 (Bethesda). 2012. PMID: 22670226 Free PMC article. - Histone Chaperones Spt6 and FACT: Similarities and Differences in Modes of Action at Transcribed Genes.
Duina AA. Duina AA. Genet Res Int. 2011;2011:625210. doi: 10.4061/2011/625210. Epub 2011 Oct 26. Genet Res Int. 2011. PMID: 22567361 Free PMC article. - Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast.
Hyle JW, Shaw RJ, Reines D. Hyle JW, et al. J Biol Chem. 2003 Aug 1;278(31):28470-8. doi: 10.1074/jbc.M303736200. Epub 2003 May 13. J Biol Chem. 2003. PMID: 12746440 Free PMC article. - Yeast PAF1 complex counters the pol III accumulation and replication stress on the tRNA genes.
Bhalla P, Shukla A, Vernekar DV, Arimbasseri AG, Sandhu KS, Bhargava P. Bhalla P, et al. Sci Rep. 2019 Sep 9;9(1):12892. doi: 10.1038/s41598-019-49316-5. Sci Rep. 2019. PMID: 31501524 Free PMC article.
References
- Archambault J., Chambers,R.S., Kobor,M.S., Ho,Y., Cartier,M., Bolotin,D., Andrews,B., Kane,C.M. and Greenblatt,J. (1997) An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 14300–14305. - PMC - PubMed
- Bortvin A. and Winston,F. (1996) Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science, 272, 1473–1476. - PubMed
- Brewster N.K., Johnston,G.C. and Singer,R.A. (1998) Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem., 273, 21972–21979. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM 60479/GM/NIGMS NIH HHS/United States
- K02 AI001816/AI/NIAID NIH HHS/United States
- R01 GM060479/GM/NIGMS NIH HHS/United States
- 5P30 CA 68485-05/CA/NCI NIH HHS/United States
- AI 01816/AI/NIAID NIH HHS/United States
- R01 GM052593/GM/NIGMS NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- GM 52593/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials