Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115 - PubMed (original) (raw)
Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115
James Shorter et al. J Cell Biol. 2002.
Abstract
p115 tethers coat protein (COP)I vesicles to Golgi membranes. The acidic COOH-terminal domain of p115 links the Golgins, Giantin on COPI vesicles, to GM130 on Golgi membranes. We now show that a SNARE motif-related domain within p115 stimulates the specific assembly of endogenous Golgi SNAREpins containing the t-SNARE, syntaxin 5. p115 catalyzes the construction of a cognate GOS-28-syntaxin-5 (v-/t-SNARE) complex by first linking the SNAREs to promote their direct interaction. These events are essential for NSF-catalyzed reassembly of postmitotic Golgi vesicles and tubules into mature cisternae. Staging experiments reveal that the linking of Golgins precedes SNAREpin assembly. Thus, p115 coordinates sequential tethering and docking of COPI vesicles by first using long tethers (Golgins) and then short tethers (SNAREs).
Figures
Figure 1.
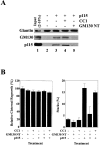
A SNARE motif-related region in p115 binds specific SNAREs. (A) The domain architecture of p115, syntaxin-5 (a t-SNARE), and GOS-28 (a v-SNARE). p115 consists of a globular head domain (H, blue), a tail domain (T) containing four coiled-coil domains (CC1-4, green), and an acidic COOH-terminal domain (A, red). SNAREs contain an ∼60 amino acid membrane proximal coiled-coil domain (green) termed the SNARE motif followed by a basic linker region (yellow) and a transmembrane domain (orange). t-SNAREs possess additional NH2-terminal coiled-coil regions. The first coiled-coil domain of p115 (CC1) displays weak homology to the SNARE motif (Weimbs et al., 1997). (B) RLGs (20 μg) were extracted with Triton X-100 buffer, clarified, and incubated with Neutravidin beads (mock) or beads bound to biotinylated p115 or CC1-4 peptides. Washed beads were eluted, and eluates were fractionated by SDS-PAGE and silver stained. Asterisks denote proteins selectively retained on p115 and CC1 beads but not others. Crosses denote proteins selectively retained on p115, CC1, and CC4 beads but not others. The triangle denotes p115 eluted from p115 beads. Squares denote proteins that correspond in size to Giantin (□, top) and GM130 (□, bottom), which are retained only on p115 beads. Arrows indicate Neutravidin breakdown products. (C) Immunoblot analyses of B. In addition, His-TA and His-T (0.5 μM) were incubated with clarified Golgi detergent extract, retrieved with Ni-NTA agarose, and processed as in B before immunoblot analysis.
Figure 1.
A SNARE motif-related region in p115 binds specific SNAREs. (A) The domain architecture of p115, syntaxin-5 (a t-SNARE), and GOS-28 (a v-SNARE). p115 consists of a globular head domain (H, blue), a tail domain (T) containing four coiled-coil domains (CC1-4, green), and an acidic COOH-terminal domain (A, red). SNAREs contain an ∼60 amino acid membrane proximal coiled-coil domain (green) termed the SNARE motif followed by a basic linker region (yellow) and a transmembrane domain (orange). t-SNAREs possess additional NH2-terminal coiled-coil regions. The first coiled-coil domain of p115 (CC1) displays weak homology to the SNARE motif (Weimbs et al., 1997). (B) RLGs (20 μg) were extracted with Triton X-100 buffer, clarified, and incubated with Neutravidin beads (mock) or beads bound to biotinylated p115 or CC1-4 peptides. Washed beads were eluted, and eluates were fractionated by SDS-PAGE and silver stained. Asterisks denote proteins selectively retained on p115 and CC1 beads but not others. Crosses denote proteins selectively retained on p115, CC1, and CC4 beads but not others. The triangle denotes p115 eluted from p115 beads. Squares denote proteins that correspond in size to Giantin (□, top) and GM130 (□, bottom), which are retained only on p115 beads. Arrows indicate Neutravidin breakdown products. (C) Immunoblot analyses of B. In addition, His-TA and His-T (0.5 μM) were incubated with clarified Golgi detergent extract, retrieved with Ni-NTA agarose, and processed as in B before immunoblot analysis.
Figure 1.
A SNARE motif-related region in p115 binds specific SNAREs. (A) The domain architecture of p115, syntaxin-5 (a t-SNARE), and GOS-28 (a v-SNARE). p115 consists of a globular head domain (H, blue), a tail domain (T) containing four coiled-coil domains (CC1-4, green), and an acidic COOH-terminal domain (A, red). SNAREs contain an ∼60 amino acid membrane proximal coiled-coil domain (green) termed the SNARE motif followed by a basic linker region (yellow) and a transmembrane domain (orange). t-SNAREs possess additional NH2-terminal coiled-coil regions. The first coiled-coil domain of p115 (CC1) displays weak homology to the SNARE motif (Weimbs et al., 1997). (B) RLGs (20 μg) were extracted with Triton X-100 buffer, clarified, and incubated with Neutravidin beads (mock) or beads bound to biotinylated p115 or CC1-4 peptides. Washed beads were eluted, and eluates were fractionated by SDS-PAGE and silver stained. Asterisks denote proteins selectively retained on p115 and CC1 beads but not others. Crosses denote proteins selectively retained on p115, CC1, and CC4 beads but not others. The triangle denotes p115 eluted from p115 beads. Squares denote proteins that correspond in size to Giantin (□, top) and GM130 (□, bottom), which are retained only on p115 beads. Arrows indicate Neutravidin breakdown products. (C) Immunoblot analyses of B. In addition, His-TA and His-T (0.5 μM) were incubated with clarified Golgi detergent extract, retrieved with Ni-NTA agarose, and processed as in B before immunoblot analysis.
Figure 2.
p115 stimulates specific SNARE complex formation in Golgi extracts. (A and B) Salt-washed RLGs that had been treated with NSF to disassemble cis-SNARE complexes were solubilized in Triton X-100 buffer and incubated with increasing concentrations of p115 (0–100 nM). GOS-28 (A) or syntaxin-5 (B) was immunoprecipitated, and the extent of coprecipitation of other Golgi SNAREs and tethers was determined by immunoblot. For the p115 immunoblot, the input (10%) lane reflects 10% of the maximum p115 concentration added (100 nM). (C and D) Quantitation of GOS-28 (C) and syntaxin-5 (D) immunoprecipitations. Amount of coprecipitated SNARE retained (% of total) as determined by densitometric scanning is plotted versus p115 (nM). Values represent means ± SEM (n = 3). (E and F) Golgi detergent extract (as in A) was incubated on ice with or without 100 nM p115 plus either buffer, 10 μM CC1 or CC2, 20 μM p115 CT (p115 COOH-terminal 75 aa), or GM130 NT (GM130 NH2-terminal 73 aa). GOS-28 (E) or syntaxin-5 (F) was immunoprecipitated, and the extent of coprecipitation of other Golgi SNAREs and p115 was determined by immunoblot. For the p115 immunoblot, the input (10%) lane reflects 10% of the maximum p115 concentration added (100 nM). (G) Salt-washed NSF-treated RLGs were incubated for 30 min at 37°C with or without 100 nM p115 plus or minus 10 μM CC1 or CC2. Reactions were stopped with SDS-PAGE sample buffer, incubated for 7 min at 25 or 95°C, and processed for immunoblot. Blots were probed with a mixture of anti–GOS-28 and anti–syntaxin-5 antibodies. Asterisk denotes a p115-induced high molecular weight species.
Figure 3.
p115 stimulates assembly of SNAREpins containing either GS15–Ykt6p–GOS-28–syntaxin-5 or membrin–Bet1p–rSec22p–syntaxin-5. (A) Salt-washed RLGs that had been treated with NSF–α-SNAP to disassemble cis-SNARE complexes were solubilized in Triton X-100 buffer and incubated for 0 (lane 2) or 60 min (lanes 3–14) on ice with increasing concentrations of p115 (0–100 nM) in the presence or absence of 10 μM CC1 or CC2. GS15 was immunoprecipitated, and retrieved beads were then either washed with Triton X-100 buffer (containing 150 mM KCl; lanes 2–11) or Triton X-100 buffer supplemented with 10 μM CC1 (lane 12), 10 μM CC2 (lane 13), or 1 M KCl (lane 14). Beads were eluted with SDS-PAGE sample buffer, and the extent of coprecipitation of other Golgi SNAREs and p115 was determined by immunoblot. For the p115 immunoblot, the input (10%) lane reflects 10% of the maximum p115 concentration added (100 nM). (B) SDS resistance of retrieved immunocomplexes. Experiments were performed as in A (using 100 nM p115) except that at the end of the immunoprecipitation beads were eluted with SDS-PAGE sample buffer and incubated at either 25 (lanes 1, 3, 5, and 7) or 95°C for 7 min (lanes 2, 4, 6, and 8). Each panel reflects a different strip of nitrocellulose probed with a different antibody (which is denoted on the far right). (C and D) Reactions were performed as in A and B except that membrin was immunoprecipitated.
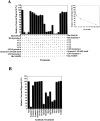
Figure 4.
The SNARE motif-related region of p115 inhibits NSF-catalyzed Golgi reassembly. (A) MGFs were incubated at 37°C for 1 h with NSF, SNAPs, and p115 (130 nM) in the presence or absence of 13 μM of the indicated SNARE or peptide. CC2-4 peptides were also added in combination. CC1-4 were also preincubated with an equimolar amount of His–GOS-28 or His–syntaxin-5 and then added. Reactions were terminated by fixation, processed for EM, and the amount of relative cisternal regrowth was determined. 100% relative cisternal regrowth represents an increase from 25 to 75% of the total membrane present as cisternae. Values represent means ± SEM (n = 3–6). (Inset) Increasing amounts of CC1 were added to the NSF reaction. Values represent means ± SEM (n = 3). (B) MGFs were incubated at 37°C for 1 h with NSF, SNAPs, and p115 with or without various anti-SNARE antibodies. Reactions were processed as in A. Values represent means ± SEM (n = 3).
Figure 5.
The SNARE motif-related domain of p115 does not disrupt Giantin-p115-GM130 tethers. (A) RLGs were dissolved in Triton X-100 buffer and incubated for 1 h on ice in either the presence or absence of p115 (250 nM) plus or minus 50 μM CC1 or GM130 NT. Giantin was then immunoprecipitated, and coprecipitation of GM130 and p115 was determined by immunoblot. The input lane reflects 2% total p115 input and 10% total GM130 and Giantin input. (B) MGFs were incubated at 37°C for 1 h with p97/p47 plus or minus p115 (130 nM). 13 μM CC1 or GM130 NT was added to some reactions. Reactions were terminated by fixation, processed for EM, and the amount of relative cisternal regrowth was determined. 100% relative cisternal regrowth represents an increase from 25 to 75% of the total membrane present as cisternae. The percentage total membrane present as stacks was also determined. Values represent means ± SEM (n = 2).
Figure 6.
p115 binds GOS-28 and syntaxin-5 directly. (A) His–syntaxin-5 (0.4 μM; top) or His–GOS-28 (0.5 μM; bottom) was incubated for 1 h on ice with p115 (0.13 μM) plus or minus 13 μM CC1, CC2, or CC3. SNAREs were retrieved with Ni-NTA agarose. p115 alone was the control. Bound proteins were processed for immunoblot. (B) p115, H, TA, or T (0.38 μM) was incubated for 1 h on ice with either His–GOS-28, His-VAMP2, GST–syntaxin-5, or GST–syntaxin-1 (20 nM). His–GOS-28 and His-VAMP2 were then immunoprecipitated with specific antibodies. GST–syntaxin-5 and GST–syntaxin-1 were retrieved with glutathione-sepharose and processed as in A. (C and D) GST–syntaxin-5 (12 pmol; C) or His–GOS-28 (12 pmol; D) was incubated for 1 h on ice with increasing concentrations of p115 (0–20 μM) plus or minus CC1 (13 μM). SNAREs were retrieved, and bound proteins were processed for immunoblot. The amount of p115 bound was determined by densitometry. Datapoints represent means (n = 3). Binding isotherms were fitted to obtain apparent K d estimates.
Figure 7.
p115 stimulates complex formation between His–GOS-28 and GST–syntaxin-5. (A) GST–syntaxin-5 (75 nM) was incubated for 1 h on ice with His–GOS-28 (75 nM) and increasing concentrations of p115 (0–375 nM). GST–syntaxin-5 (top) or His–GOS-28 (bottom) was retrieved. Controls omitted the SNARE to be retrieved plus or minus p115. Bound proteins were processed for immunoblot. For the p115 immunoblot, the input (10%) lane reflects 10% of the maximum p115 concentration added (375 nM). (B) GST–syntaxin-5 (75 nM) was incubated for 1 h on ice with His–GOS-28 (75 nM) in the presence or absence of p115 (150 nM) with or without 15 μM CC1 or CC2, 30 μM p115 CT, or GM130 NT. GST–syntaxin-5 (top) or His–GOS-28 (bottom) was retrieved. Controls omitted the SNARE to be retrieved plus or minus p115. Reactions were processed as in A. Note the longer exposure time required to detect the coprecipitated SNARE (His–GOS-28, top, and GST–syntaxin-5, bottom). (C and D) GST–syntaxin-5 (12 pmol) was incubated for 1 h on ice with increasing concentrations of His–GOS-28 (0–20 μM) in the presence or absence of CC1 (20 μM), plus (D) or minus (C) p115 (0.2 μM). GST–syntaxin-5 was retrieved. Reactions were processed as in A. The amount of His–GOS-28 bound was determined by densitometry. Datapoints represent means (n = 3), and binding isotherms were fitted. (E) Reactions were performed as in A except p115 was replaced with His-H, His-TA, or His-T (0–3 μM).
Figure 7.
p115 stimulates complex formation between His–GOS-28 and GST–syntaxin-5. (A) GST–syntaxin-5 (75 nM) was incubated for 1 h on ice with His–GOS-28 (75 nM) and increasing concentrations of p115 (0–375 nM). GST–syntaxin-5 (top) or His–GOS-28 (bottom) was retrieved. Controls omitted the SNARE to be retrieved plus or minus p115. Bound proteins were processed for immunoblot. For the p115 immunoblot, the input (10%) lane reflects 10% of the maximum p115 concentration added (375 nM). (B) GST–syntaxin-5 (75 nM) was incubated for 1 h on ice with His–GOS-28 (75 nM) in the presence or absence of p115 (150 nM) with or without 15 μM CC1 or CC2, 30 μM p115 CT, or GM130 NT. GST–syntaxin-5 (top) or His–GOS-28 (bottom) was retrieved. Controls omitted the SNARE to be retrieved plus or minus p115. Reactions were processed as in A. Note the longer exposure time required to detect the coprecipitated SNARE (His–GOS-28, top, and GST–syntaxin-5, bottom). (C and D) GST–syntaxin-5 (12 pmol) was incubated for 1 h on ice with increasing concentrations of His–GOS-28 (0–20 μM) in the presence or absence of CC1 (20 μM), plus (D) or minus (C) p115 (0.2 μM). GST–syntaxin-5 was retrieved. Reactions were processed as in A. The amount of His–GOS-28 bound was determined by densitometry. Datapoints represent means (n = 3), and binding isotherms were fitted. (E) Reactions were performed as in A except p115 was replaced with His-H, His-TA, or His-T (0–3 μM).
Figure 8.
p115 catalyzes SNARE assembly and is not required to maintain SNARE complexes. (A) GST–syntaxin-5 (75 nM) was incubated for 1 h on ice with His–GOS-28 (75 nM) and p115 (0–150 nM). GST–syntaxin-5 was retrieved. His–GOS-28 alone or plus p115 served as controls. Beads were washed with either buffer containing 150 mM KCl, 1 M KCl, 20 μM CC1, or CC2. Bound proteins were processed for immunoblot. (B) GST–syntaxin-5 (75 nM) was incubated on ice with His–GOS-28 (75 nM) and p115 (150 nM). At various times during the incubation, the reaction was supplemented with buffer, 15 μM CC1, or CC2. After 1 h, GST–syntaxin-5 was retrieved, and bound proteins were processed for immunoblot. (C) GST–syntaxin-5 (75 nM) was incubated with His–GOS-28 (75 nM) and p115 (75 pM) for 4 h at 4°C with agitation. Reactions were processed as in B. (D) GST–syntaxin-5 (75 nM) was bound to glutathione beads and incubated with His–GOS-28 (75 nM) plus or minus p115 (75 pM) for various times ranging from 2 min to 18 h. Beads were recovered, and the amount of His–GOS-28 binding was determined by immunoblot. (E) Salt-washed RLGs that had been treated with NSF–α-SNAP to disassemble cis-SNARE complexes were solubilized in Triton X-100 buffer and incubated for 4 h on ice plus or minus p115 (100 pM). GS15 was immunoprecipitated, and retrieved beads were then washed with Triton X-100 buffer. Beads were eluted with SDS-PAGE sample buffer, and the extent of coprecipitation of other Golgi SNAREs and p115 was determined by immunoblot. (F) Reactions were performed as in E in the presence or absence of p115 (100 pM), and the incubation time varied from 30 min to 18 h.
Figure 9.
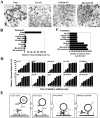
Resolution of vesicle tethering and SNARE assembly during NSF-driven Golgi reassembly. (A) Morphology of NSF reaction products in the presence of various inhibitors. Note the presence of stacks of cisternae in the absence of inhibitor compared with dispersed tubules and vesicles in the MGFs (ice + fix) or in the presence of GM130 NT, and the highly clustered tubules and vesicles in the presence of His–GOS-28. Bar, 0.5 μm. (B) MGFs were incubated at 37°C for 1 h with NSF, SNAPs, and p115 (0.13 μM). Various inhibitors were added: GM130 NT (13 μM), Gtn1-448 (13 μM), GDI (14 μM), chrysin (120 μM), CC1 (13 μM), His–GOS-28 (6 μM), or His–syntaxin-5 (6 μM). Dilution (10×) was with reaction buffer containing NSF, SNAPs, and p115. Reactions were terminated by fixation, processed for EM, and the amount of relative cisternal regrowth was determined. 100% relative cisternal regrowth represents an increase from 25 to 75% of the total membrane present as cisternae. Values represent means ± SEM (n = 3). (C) Quantitation of density of tubules/vesicles per μm2 for the reactions described in B. (D) Kinetic sensitivity of NSF reaction to various inhibitors. NSF reactions were performed as in B except that at the indicated times reactions were either stopped by fixation or treated as indicated and incubated for a total of 1 h at 37°C. Reactions were processed as in B. Values represent means ± SEM (n = 4). (E) A model depicting the proposed sequence of events during GOS-28–syntaxin-5 complex assembly in NSF-driven Golgi reassembly (see Discussion).
Comment in
- Vesicle tethers promoting fusion machinery assembly.
Söllner TH. Söllner TH. Dev Cell. 2002 Apr;2(4):377-8. doi: 10.1016/s1534-5807(02)00161-2. Dev Cell. 2002. PMID: 11970884 Review.
Similar articles
- Phosphorylation of the vesicle-tethering protein p115 by a casein kinase II-like enzyme is required for Golgi reassembly from isolated mitotic fragments.
Dirac-Svejstrup AB, Shorter J, Waters MG, Warren G. Dirac-Svejstrup AB, et al. J Cell Biol. 2000 Aug 7;150(3):475-88. doi: 10.1083/jcb.150.3.475. J Cell Biol. 2000. PMID: 10931861 Free PMC article. - p115-SNARE interactions: a dynamic cycle of p115 binding monomeric SNARE motifs and releasing assembled bundles.
Wang T, Grabski R, Sztul E, Hay JC. Wang T, et al. Traffic. 2015 Feb;16(2):148-71. doi: 10.1111/tra.12242. Epub 2015 Jan 4. Traffic. 2015. PMID: 25406594 Free PMC article. - A role for giantin in docking COPI vesicles to Golgi membranes.
Sönnichsen B, Lowe M, Levine T, Jämsä E, Dirac-Svejstrup B, Warren G. Sönnichsen B, et al. J Cell Biol. 1998 Mar 9;140(5):1013-21. doi: 10.1083/jcb.140.5.1013. J Cell Biol. 1998. PMID: 9490716 Free PMC article. - SNAREs and membrane fusion in the Golgi apparatus.
Nichols BJ, Pelham HR. Nichols BJ, et al. Biochim Biophys Acta. 1998 Aug 14;1404(1-2):9-31. doi: 10.1016/s0167-4889(98)00044-5. Biochim Biophys Acta. 1998. PMID: 9714710 Review. - Vesicle tethers promoting fusion machinery assembly.
Söllner TH. Söllner TH. Dev Cell. 2002 Apr;2(4):377-8. doi: 10.1016/s1534-5807(02)00161-2. Dev Cell. 2002. PMID: 11970884 Review.
Cited by
- Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2).
Hammond GR, Schiavo G, Irvine RF. Hammond GR, et al. Biochem J. 2009 Jul 29;422(1):23-35. doi: 10.1042/BJ20090428. Biochem J. 2009. PMID: 19508231 Free PMC article. - Membrane fusion.
Wickner W, Schekman R. Wickner W, et al. Nat Struct Mol Biol. 2008 Jul;15(7):658-64. doi: 10.1038/nsmb.1451. Nat Struct Mol Biol. 2008. PMID: 18618939 Free PMC article. Review. - An active tethering mechanism controls the fate of vesicles.
An SJ, Rivera-Molina F, Anneken A, Xi Z, McNellis B, Polejaev VI, Toomre D. An SJ, et al. Nat Commun. 2021 Sep 14;12(1):5434. doi: 10.1038/s41467-021-25465-y. Nat Commun. 2021. PMID: 34521845 Free PMC article. - Glycosyltransferase-specific Golgi-targeting mechanisms.
Petrosyan A, Ali MF, Cheng PW. Petrosyan A, et al. J Biol Chem. 2012 Nov 2;287(45):37621-7. doi: 10.1074/jbc.C112.403006. Epub 2012 Sep 17. J Biol Chem. 2012. PMID: 22988244 Free PMC article. - News and Views into the SNARE Complexity in Arabidopsis.
Kim SJ, Brandizzi F. Kim SJ, et al. Front Plant Sci. 2012 Feb 10;3:28. doi: 10.3389/fpls.2012.00028. eCollection 2012. Front Plant Sci. 2012. PMID: 23018380 Free PMC article.
References
- Allan, B.B., B.D. Moyer, and W.E. Balch. 2000. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 289:444–448. - PubMed
- Baker, D., and D.A. Agard. 1994. Kinetics versus thermodynamics in protein folding. Biochemistry. 33:7505–7509. - PubMed
- Barrick, D., and F.M. Hughson. 2002. Irreversible assembly of membrane fusion machines. Nat. Struct. Biol. 9:78–80. - PubMed
- Chen, Y.A., S.J. Scales, S.M. Patel, Y.C. Doung, and R.H. Scheller. 1999. SNARE complex formation is triggered by Ca2+ and drives membrane fusion. Cell. 97:165–174. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases