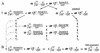Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster - PubMed (original) (raw)
Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster
Mikhail Savitsky et al. Mol Cell Biol. 2002 May.
Abstract
Telomeres of Drosophila melanogaster contain arrays of the retrotransposon-like elements HeT-A and TART. Their transposition to broken chromosome ends has been implicated in chromosome healing and telomere elongation. We have developed a genetic system which enables the determination of the frequency of telomere elongation events and their mechanism. The frequency differs among lines with different genotypes, suggesting that several genes are in control. Here we show that the Su(var)2-5 gene encoding heterochromatin protein 1 (HP1) is involved in regulation of telomere length. Different Su(var)2-5 mutations in the heterozygous state increase the frequency of HeT-A and TART attachment to the broken chromosome end by more than a hundred times. The attachment occurs through either HeT-A/TART transposition or recombination with other telomeres. Terminal DNA elongation by gene conversion is greatly enhanced by Su(var)2-5 mutations only if the template for DNA synthesis is on the same chromosome but not on the homologous chromosome. The Drosophila lines bearing the Su(var)2-5 mutations maintain extremely long telomeres consisting of HeT-A and TART for many generations. Thus, HP1 plays an important role in the control of telomere elongation in D. melanogaster.
Figures
FIG. 1.
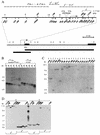
Molecular structure of the broken chromosome end in the yellow gene. (A) A schematic presentation of terminal yellow deficiencies associated with different y phenotypes. The promoter (ATATAAAA) and start of translation (ATG) locations are indicated relative to the transcription start site of the yellow gene. The coding yellow region is shown as a black box. The location of the start of the yellow transcription region is shown by a horizontal arrow on the uppermost solid horizontal line. The dotted horizontal lines show the regions of the yellow sequence in which the termini of the yTD line that correspond to the same classes of y phenotype have been mapped. The _Kpn_I-_Eco_RV and _Bam_HI-_Eco_RI genomic fragments used as a probe for Southern blot analysis are indicated by the bottommost thick horizontal line and the line just above, respectively. The points of HeT-A attachment are shown by small vertical arrows below the uppermost solid horizontal line. The points of TART attachment are shown by small vertical arrows above the same line. Restriction enzyme abbreviations: B, _Bam_HI; K, _Kpn_I; H, _Hin_dIII; E, _Eco_RV; N, _Nru_I; S, _Spe_I; R, _Eco_RI. (B) Effect of the _Su(var)2_-505 mutation on the rate of terminal DNA shortening. Southern blot analysis of DNAs prepared from 10 to 14 yTD/yac females of the _Su(var)2_-505 and control lines taken in six subsequent generations as described in the Fig. 2A legend. DNAs were digested with _Eco_RI. The filter was hybridized with the _Bam_HI-_Eco_RI probe. The additional bands correspond to new DNA attachments to the receding yellow sequences at the end of truncated chromosome. (C) HeT-A and TART transpositions to the broken chromosome end in the yellow gene. DNAs were digested with _Eco_RV (E), _Nru_I (N), and _Spe_I (S). The filter was hybridized with the _Kpn_I-_Eco_RV probe. (D) HeT-A and TART transpositions obtained in the progeny of one _yTD/yac; Su(var)2_-505/CyO female displaying a y1 phenotype. In the next generation, all yTD/yac; If/CyO daughters with either a y1- or a y2-like phenotype were individually crossed for DNA isolation. DNAs were digested with _Kpn_I (K) and _Eco_RV (E). The filter was hybridized with the _Kpn_I-_Eco_RV probe. The junctions between yellow and new DNA attachments were cloned and sequenced in the cases designated with numbers (see Fig. 3).
FIG. 2.
The scheme used to study the role of Su(var) mutations in terminal DNA elongation. (A) The main scheme of crosses. The mutations of _Su(var)2_-5 tested were _Su(var)2_-501, _Su(var)2_-502, _Su(var)2_-504, or _Su(var)2_-505. Depending on the purpose of the experiment, the yTD chromosome contains different y alleles at the end of the terminally truncated chromosome. In some experiments, yac chromosome (yac*) was substituted for either the yw or the ytataw chromosome. (B) The scheme for isolation of individual stable lines bearing terminally truncated chromosomes with new DNA attachments. yTD* indicates a y allele with a new phenotype (i.e., a new DNA attachment).
FIG. 3.
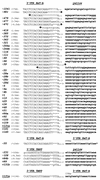
Diagram of HeT-A and TART attachment to the receding yellow sequences. All HeT-A and TART additions are indicated as described in the legends to Fig. 1, 4, and 5. The numbers in parentheses show the approximate sizes of the attachments. The base pair locations at the junctions between HeT-A or TART and yellow are shown. The lowercase letters indicate substitutions in the conserved sequences at the 3′ end of HeT-A or TART. Adenine bases at the junctions may originate either from the yellow sequences or from the 3′ oligo(A) tail of HeT-A or TART.
FIG. 4.
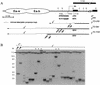
The model system to study terminal DNA elongation by gene conversion on the _Su(var)2_-5 mutant background in the presence of a template on the homologous chromosome. (A) Schematic presentation of the terminal yellow deficiencies associated with different y alleles. The molecular structures of the y1 and ytata mutations are shown. The wing (En-w) and body (En-b) enhancers are indicated by ovoids. The approximate regions of the ends of terminally truncated chromosomes in the yTD alleles are shown by three thin horizontal black lines. The dotted horizontal lines show the regions of yellow sequence in which the termini of yTD line with original phenotype have been mapped. The dashed horizontal lines show the regions of yellow sequence in which the termini of yTD line acquiring a new y phenotype have been mapped. The 3.2-kb _Bam_HI-_Eco_RI genomic fragment used as a probe for Southern blot analysis is indicated by the uppermost thick line. The HeT-A attachment points are shown by small vertical arrows below each of the three thin horizontal black lines. The sequences at the junctions between yellow and HeT-A are shown in Fig. 3. Other designations are as defined in the Fig. 1 legend. (B) Southern blot analysis of DNAs prepared from F2 progeny of individual yTD+250/ytataw; _Su(var)2_-5/CyO flies. DNAs were digested with _Eco_RI. The filter was hybridized with the 3.2-kb _Bam_HI-_Eco_RI probe. The 17.3-kb band corresponds to the DNA fragment between two _Eco_RI sites in the ytata allele. Asterisks indicate yTD lines displaying a y1-like phenotype that acquired new DNA attachments. The presence of additional bands indicates the heterogeneity of the progeny, suggesting that in some sisters, terminally truncated chromosomes acquired new DNA sequences (HeT-A or TART).
FIG. 5.
Model system to study terminal DNA elongation by gene conversion on a _Su(var)2_-5 mutant background in the presence of a template on the same chromosome. (A) Schematic presentation of the yTD2h2 allele and its derivatives associated with different y phenotypes. The approximate end of the truncated chromosome in the yTD2h2 allele is shown by a downward-pointing vertical arrow. The coordinates (kilobases) in the yellow gene and gypsy element are defined from the transcription start site of the distal yellow promoter. The gypsy element is inserted −700 bp upstream of the transcription start site. The Su(Hw) binding sites are indicated by the stripes in the striped boxes. The wing (En-w) and body (En-b) enhancers are indicated by ovoids. The triangle and arrowhead indicate the hobo element and its direction, respectively. Abbreviations: d-pr, distal yellow promoter; p-pr, proximal yellow promoter; d-Su(Hw), distal gypsy insulator; p-Su(Hw), proximal gypsy insulator; d-gypsy, distal gypsy retrotransposon; p-gypsy, proximal gypsy retrotransposon. The approximate ends of the truncated chromosomes in two yTD2h2 derivatives are shown by the bottommost two thin black lines. The dotted horizontal lines show the regions of the yellow sequence in which the termini of the yTD line with the y2-like phenotype have been mapped. The dashed horizontal lines show the regions of the yellow sequence where the termini of yTD line acquiring a yr (yellow revertant)-like phenotype have been mapped. The _Hin_dIII-_Bam_HI genomic fragment used as a probe for Southern blot analysis is indicated by thick line segments. The points of HeT-A attachment are shown by small arrows with numbers. The sequences at the junctions between yellow and HeT-A are shown in Fig. 3. Restriction enzyme abbreviations: B, _Bam_HI; K, _Kpn_I; H, _Hin_dIII; N, _Nco_I; G, _Bgl_II; X, _Xho_I. Other designations are defined in the Fig. 1 legend. (B) Diagram of HeT-A attachment to the receding yellow or gypsy sequences. All HeT-A additions are indicated as in Fig. 5A. Other designations are defined in the Fig. 3 legend. (C) Southern blot analysis of DNAs prepared from F2 progeny of individual yTD2h2/yac; _Su(var)2_-5/CyO02 flies. DNAs were digested with _Bam_HI. The filter was hybridized with the _Hin_dIII-_Bam_HI probe. The 13-kb band corresponds to the DNA fragment that hybridized with the proximal _Hin_dIII-_Bam_HI probe. The presence of additional bands indicates the heterogeneity of the progeny, suggesting that in some sisters, terminally truncated chromosomes acquired new DNA sequences.
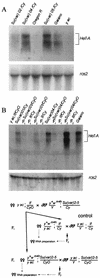
FIG. 6.
Analysis of HeT-A transcripts in the _Su(var)2_-5 mutant strains. (A) Northern blot hybridization with total RNA isolated from progeny from the In(I)wm4h; Su(var)2-504/In(2L)tIn(2R)Cy[Su(var)04/Cy], Df(1)w, and y1w67c23; Su(var)2-502/CyO y+[Su(var)02/Cy]; Oregon R lines and the In(I)wm4h; Su(var)2-505/In(2L)Cy1 In(2R)Cy[Su(var)05/Cy]; Gaiano and yac lines. The double-strand probe from the open reading frame of HeT-A detects a prominent RNA of ∼6 kb. The probe from the ras2 gene gives rise to a 1.6-kb transcript that is used as a marker for the amount of RNA. (B) Northern blot hybridization with total RNA isolated from flies obtained in the crosses is depicted in the lower part of the figure. Gaiano and If/CyO lines were used as controls with high and low HeT-A transcription, respectively. The In(I)wm4h; Su(var)2-501/SM1 [Su(var)01/Cy] and In(I)wm4h; Su(var)2-504/In(2L)tIn(2R)Cy Cy1Roi1pR1cn1[Su(var)04/Cy] lines were used in the crosses.
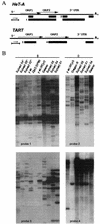
FIG. 7.
The content of HeT-A and TART elements in the _Su(var)2_-5 mutants. (A) Diagrams of HeT-A and TART. The bars under each diagram indicate the sequences used as probes for Southern blot analysis. (B) Southern blot analysis of HeT-A and TART copy numbers in the _Su(var)2_-5 lines. Asterisks indicate new lines obtained from J. Eissenberg (38) as follows: Su(var)02*, Df(1)w y1w67c23; Su(var)2-502/CyO y+; Su(var)04*, Df(1)w y1w67c23; Su(var)2-504/CyO y+; Su(var)05*, and Df(1)w y1w67c23; Su(var)2-505/CyO y+. The controls were Gaiano (very long telomeres), yac; If/CyO, and Oregon R (normal telomeres). Results from progeny from the Su(var)01, If/CyO In(I)wm4h; Su(var)2-501/SM; Su(var)04, Df(1)w y1w67c23; Su(var)2-504/CyO y+; Su(var)05, In(I)wm4h; Su(var)2-505/In(2L)Cy1 In(2R)Cy, Dp(2, 2)P90 yw, and Dp (2, 2)P90/CyOy+ lines are shown. DNAs were digested with _Bam_HI (B) or _Eco_RI (R). The filters were probed with different fragments indicated in panel A.
Similar articles
- Enhancer of terminal gene conversion, a new mutation in Drosophila melanogaster that induces telomere elongation by gene conversion.
Melnikova L, Georgiev P. Melnikova L, et al. Genetics. 2002 Nov;162(3):1301-12. doi: 10.1093/genetics/162.3.1301. Genetics. 2002. PMID: 12454074 Free PMC article. - Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres.
Siriaco GM, Cenci G, Haoudi A, Champion LE, Zhou C, Gatti M, Mason JM. Siriaco GM, et al. Genetics. 2002 Jan;160(1):235-45. doi: 10.1093/genetics/160.1.235. Genetics. 2002. PMID: 11805059 Free PMC article. - Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends.
Rashkova S, Karam SE, Kellum R, Pardue ML. Rashkova S, et al. J Cell Biol. 2002 Nov 11;159(3):397-402. doi: 10.1083/jcb.200205039. Epub 2002 Nov 4. J Cell Biol. 2002. PMID: 12417578 Free PMC article. - Drosophila telomere elongation.
Biessmann H, Walter MF, Mason JM. Biessmann H, et al. Ciba Found Symp. 1997;211:53-67; discussion 67-70. doi: 10.1002/9780470515433.ch5. Ciba Found Symp. 1997. PMID: 9524751 Review. - Control of telomere elongation and telomeric silencing in Drosophila melanogaster.
Mason JM, Haoudi A, Konev AY, Kurenova E, Walter MF, Biessmann H. Mason JM, et al. Genetica. 2000;109(1-2):61-70. doi: 10.1023/a:1026548503320. Genetica. 2000. PMID: 11293796 Review.
Cited by
- Telomeric retrotransposons show propensity to form G-quadruplexes in various eukaryotic species.
Jedlička P, Tokan V, Kejnovská I, Hobza R, Kejnovský E. Jedlička P, et al. Mob DNA. 2023 Apr 10;14(1):3. doi: 10.1186/s13100-023-00291-9. Mob DNA. 2023. PMID: 37038191 Free PMC article. - Mod(mdg4) variants repress telomeric retrotransposon HeT-A by blocking subtelomeric enhancers.
Takeuchi C, Yokoshi M, Kondo S, Shibuya A, Saito K, Fukaya T, Siomi H, Iwasaki YW. Takeuchi C, et al. Nucleic Acids Res. 2022 Nov 11;50(20):11580-11599. doi: 10.1093/nar/gkac1034. Nucleic Acids Res. 2022. PMID: 36373634 Free PMC article. - Identification of the Telomere elongation Mutation in Drosophila.
Reddy HM, Randall TA, Cipressa F, Porrazzo A, Cenci G, Frydrychova RC, Mason JM. Reddy HM, et al. Cells. 2022 Nov 3;11(21):3484. doi: 10.3390/cells11213484. Cells. 2022. PMID: 36359878 Free PMC article. - Loss of telomere silencing is accompanied by dysfunction of Polo kinase and centrosomes during Drosophila oogenesis and early development.
Morgunova V, Kordyukova M, Mikhaleva EA, Butenko I, Pobeguts OV, Kalmykova A. Morgunova V, et al. PLoS One. 2021 Oct 8;16(10):e0258156. doi: 10.1371/journal.pone.0258156. eCollection 2021. PLoS One. 2021. PMID: 34624021 Free PMC article. - Unravelling HP1 functions: post-transcriptional regulation of stem cell fate.
Casale AM, Cappucci U, Piacentini L. Casale AM, et al. Chromosoma. 2021 Sep;130(2-3):103-111. doi: 10.1007/s00412-021-00760-1. Epub 2021 Jun 15. Chromosoma. 2021. PMID: 34128099 Free PMC article. Review.
References
- Ashburner, M. 1989. Drosophila: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- Biessmann, H., B. Kasravi, T. Bui, G. Fujiwara, L. E. Champion, and J. M. Mason. 1994. Comparison of two active HeT-A retroposons of Drosophila melanogaster. Chromosoma 103:90-98. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials