Nup98 is a mobile nucleoporin with transcription-dependent dynamics - PubMed (original) (raw)
Nup98 is a mobile nucleoporin with transcription-dependent dynamics
Eric R Griffis et al. Mol Biol Cell. 2002 Apr.
Abstract
Nucleoporin 98 (Nup98), a glycine-leucine-phenylalanine-glycine (GLFG) amino acid repeat-containing nucleoporin, plays a critical part in nuclear trafficking. Injection of antibodies to Nup98 into the nucleus blocks the export of most RNAs. Nup98 contains binding sites for several transport factors; however, the mechanism by which this nucleoporin functions has remained unclear. Multiple subcellular localizations have been suggested for Nup98. Here we show that Nup98 is indeed found both at the nuclear pore complex and within the nucleus. Inside the nucleus, Nup98 associates with a novel nuclear structure that we term the GLFG body because the GLFG domain of Nup98 is required for targeting to this structure. Photobleaching of green fluorescent protein-Nup98 in living cells reveals that Nup98 is mobile and moves between these different localizations. The rate of recovery after photobleaching indicates that Nup98 interacts with other, less mobile, components in the nucleoplasm. Strikingly, given the previous link to nuclear export, the mobility of Nup98 within the nucleus and at the pore is dependent on ongoing transcription by RNA polymerases I and II. These data give rise to a model in which Nup98 aids in direction of RNAs to the nuclear pore and provide the first potential mechanism for the role of a mobile nucleoporin.
Figures
Figure 1
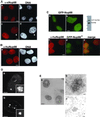
Nup98 shows a substantial intranuclear localization in human and Xenopus cells. (A) Immunofluorescence using antibodies raised against xNup98 revealed brightly staining structures as well as a diffuse intranuclear stain at interphase in XL177 cells (a). At prophase, a population of Nup98 at the nuclear rim is revealed (c, prophase cell indicated by arrowhead). b and d are stained with Hoechst. (B) Endogenous human Nup98 in live HeLa cells generates a nuclear rim stain with intranuclear foci and diffuse nucleoplasmic signal (a). b is stained with Hoechst. (C) The same localization pattern was observed in HeLa cells expressing GFP-Nup98. a is a section through the midplane of the nucleus, and b is focused on the surface. To the right is an immunoblot of transfected cells probed with antibodies raised against human Nup98. Molecular mass markers correspond to 184, 121, 86, and 68 kDa. In the bottom panels, GFP-Nup98* (d) is localized to the same intranuclear foci as the endogenous protein (c). Note that to avoid cross-reactivity with the antibody, this construct contains only the GLFG domain of Nup98, and thus signals for targeting to the nuclear pore are absent in this GFP protein. (D) Deconvolution microscopy of the endogenous xNup98 in XL177 cells shows that the intranuclear structures are clusters of smaller bodies (a and inset). The GFP-Nup98 bodies in HeLa cells appear similarly lobular (b and inset). (E) In XL177 cells, endogenous Nup98 localized using DAB and visualized by phase contrast (a) recapitulates the Nup98 localization pattern seen with immunofluorescence. Transmission electron microscopy analysis using either DAB (b) or immunogold (c) confirms that the bodies seen at the light level are in fact clusters of smaller structures. Scale bars, 5 μm in light micrographs and 100 nm in electron micrographs.
Figure 2
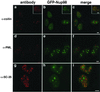
The intranuclear Nup98 bodies are novel structures. HeLa cells expressing GFP-Nup98 were stained for markers of other, previously characterized nuclear bodies. GFP-Nup98–expressing cells (b) were stained with anti-p80 coilin (a), and the two images were merged in c. The insets show the same staining in Xenopus cells. GFP-Nup98–expressing cells (e) were stained to visualize PML bodies using either mAb 5E10 (d) or an antibody to SUMO (Griffis and Powers, unpublished results), and the images were merged in f. Splicing factor speckles were visualized with mAb SC-35 (g) in GFP-Nup98–expressing cells (h), and the results are shown merged in i. Scale bars, 5 μm.
Figure 3
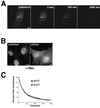
Nup98 is mobile within the nucleoplasm and dynamically associates with both the nuclear bodies and the nuclear pore complex. (A) In the top row, the indicated area containing a single nuclear body was photobleached and then imaged every 1.5 s for 2 min. To improve the time resolution for analyzing Nup98 recovery in the nucleoplasm, the inset area indicated in the first panel of the middle row was enlarged and imaged as a region of interest. The area circled in the prebleach panel was then photobleached and imaged every 150 ms for 20 s. The bottom row represents a tangential section through the nuclear envelope. The indicated area of the nuclear rim was photobleached and then imaged every 1.5 s for a total of 200 s. Insets show close-ups of an area near the edge of the bleach zone. (B) FRAP was quantitated. Values were background corrected and adjusted for bleaching as described in MATERIALS AND METHODS. The top graph represents the average recovery of fluorescence of GFP-Nup98 and free GFP after photobleaching of the nucleoplasm of transfected cells. The bottom graph shows the recovery of GFP-Nup98 in intranuclear bodies after photobleaching. Each graph represents the average of five independent experiments. Error bars depict the SEM. (C) The focal plane was set at the surface of the nuclear envelope, and the region indicated in the first panel was photobleached. The bleached area was then imaged every 20 s for 25 min, and fluorescence recovery was determined. Scale bars, 5 μm. A, top and bottom are presented as videos in the supplementary material.
Figure 3
Nup98 is mobile within the nucleoplasm and dynamically associates with both the nuclear bodies and the nuclear pore complex. (A) In the top row, the indicated area containing a single nuclear body was photobleached and then imaged every 1.5 s for 2 min. To improve the time resolution for analyzing Nup98 recovery in the nucleoplasm, the inset area indicated in the first panel of the middle row was enlarged and imaged as a region of interest. The area circled in the prebleach panel was then photobleached and imaged every 150 ms for 20 s. The bottom row represents a tangential section through the nuclear envelope. The indicated area of the nuclear rim was photobleached and then imaged every 1.5 s for a total of 200 s. Insets show close-ups of an area near the edge of the bleach zone. (B) FRAP was quantitated. Values were background corrected and adjusted for bleaching as described in MATERIALS AND METHODS. The top graph represents the average recovery of fluorescence of GFP-Nup98 and free GFP after photobleaching of the nucleoplasm of transfected cells. The bottom graph shows the recovery of GFP-Nup98 in intranuclear bodies after photobleaching. Each graph represents the average of five independent experiments. Error bars depict the SEM. (C) The focal plane was set at the surface of the nuclear envelope, and the region indicated in the first panel was photobleached. The bleached area was then imaged every 20 s for 25 min, and fluorescence recovery was determined. Scale bars, 5 μm. A, top and bottom are presented as videos in the supplementary material.
Figure 4
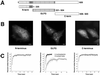
The GLFG domain of Nup98 targets to intranuclear bodies. (A) The domain organization of human Nup98 (920 amino acids) is shown. Each of the depicted domains was N terminally fused to GFP and transfected into HeLa cells. (B) The localization of each of these GFP-domain constructs in HeLa cells showed that only the GLFG domain associates with nuclear bodies. Scale bars, 5 μm. (C) FRAP analysis was carried out on each domain within the nucleoplasm following the same protocol used to quantitate the recovery of Nup98 in the nucleoplasm. The average recovery for five independent experiments is plotted (open circles). In the middle graph, the open squares represent the average recovery after photobleaching of the GFP-GLFG nuclear bodies. Note the difference in the X-axis values between GLFG and N- and C-terminal domain graphs.
Figure 5
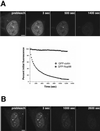
FLIP reveals that Nup98 can shuttle between the nucleus and cytoplasm. (A) An area of cytoplasm was repeatedly bleached, and the loss of fluorescence from the nucleus was measured over time. As a control, a cell expressing GFP-coilin was repeatedly bleached in the cytoplasm, and the loss of nuclear fluorescence was measured. As the graph indicates, only 8% of the nuclear GFP-coilin was photobleached over the full time course of the experiment. In contrast, GFP-Nup98 fluorescence decreased >94% during the same time course. The data points plotted are not averaged but are representative of 10 independent experiments. (B) The FLIP protocol was modified for measuring the loss of fluorescence of GFP-Nup98 at nuclear pores. The focal plane is set at the surface of the nuclear envelope and, to reduce photobleaching during imaging, fewer scans were taken over a longer time by inserting a 5-s time delay between images. Scale bar, 5 μm. A and B are included as videos in the supplementary material.
Figure 6
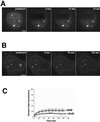
Nup98 shuttling does not require the Ran gradient. (A) tsBN2 cells were transfected with GFP-Nup98, and, before imaging, the cells were shifted to the nonpermissive temperature (39°C) for 4 h. FLIP was then carried out as described above. (B) To confirm that the temperature shift had resulted in loss of the Ran gradient, Ran was localized by immunofluorescence in tsBN2 cells either growing at 33.5°C (unshifted) or shifted to 39°C for 4 h. The shifted cells that lacked a functional RCC1 protein could not sustain the nuclear Ran gradient and thus did not maintain nuclear localization of Ran. Scale bars, 5μm. (C) Loss of GFP-Nup98 fluorescence was quantitated in both temperature-shifted and unshifted cells. The graphs are not averaged but are representative of five independent experiments. A is presented as a video in the supplementary material.
Figure 7
Nup98 mobility is sensitive to inhibitors of transcription. (A) HeLa cells expressing GFP-Nup98 were treated with actinomycin D for 4 h before FRAP analysis. The area outlined was bleached, and fluorescence recovery was measured every 1.5 s for 2 min. (B) HeLa cells expressing GFP-Nup98 were treated with DRB for 4 h before FRAP. The area outlined was bleached, and the fluorescence recovery was measured every 1.5 s for 2 min. Scale bars, 5 μm. (C) Quantitation of the recovery of Nup98 nuclear bodies shows that the mobility of Nup98 is severely reduced by transcriptional inhibitors. Each data point is the average of five independent experiments. Error bars depict the SEM. ActD, actinomycin D. A and B are presented in video form on the CD.
Similar articles
- Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility.
Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Griffis ER, et al. Mol Biol Cell. 2004 Apr;15(4):1991-2002. doi: 10.1091/mbc.e03-10-0743. Epub 2004 Jan 12. Mol Biol Cell. 2004. PMID: 14718558 Free PMC article. - Intranuclear dynamics of the Nup107-160 complex.
Morchoisne-Bolhy S, Geoffroy MC, Bouhlel IB, Alves A, Audugé N, Baudin X, Van Bortle K, Powers MA, Doye V. Morchoisne-Bolhy S, et al. Mol Biol Cell. 2015 Jun 15;26(12):2343-56. doi: 10.1091/mbc.E15-02-0060. Epub 2015 Apr 22. Mol Biol Cell. 2015. PMID: 25904327 Free PMC article. - Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export.
Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. Vasu S, et al. J Cell Biol. 2001 Oct 29;155(3):339-54. doi: 10.1083/jcb.200108007. Epub 2001 Oct 29. J Cell Biol. 2001. PMID: 11684705 Free PMC article. - Nucleoporin Nup98: a gatekeeper in the eukaryotic kingdoms.
Iwamoto M, Asakawa H, Hiraoka Y, Haraguchi T. Iwamoto M, et al. Genes Cells. 2010 Jun;15(7):661-9. doi: 10.1111/j.1365-2443.2010.01415.x. Epub 2010 Jun 7. Genes Cells. 2010. PMID: 20545767 Review. - Nucleoplasmic Nup98 controls gene expression by regulating a DExH/D-box protein.
Capitanio JS, Montpetit B, Wozniak RW. Capitanio JS, et al. Nucleus. 2018 Jan 1;9(1):1-8. doi: 10.1080/19491034.2017.1364826. Epub 2017 Sep 21. Nucleus. 2018. PMID: 28934014 Free PMC article. Review.
Cited by
- NUP98 oncofusions in myeloid malignancies: An update on molecular mechanisms and therapeutic opportunities.
Rasouli M, Troester S, Grebien F, Goemans BF, Zwaan CM, Heidenreich O. Rasouli M, et al. Hemasphere. 2024 Sep 25;8(9):e70013. doi: 10.1002/hem3.70013. eCollection 2024 Sep. Hemasphere. 2024. PMID: 39323480 Free PMC article. Review. - Disclosing the Interactome of Leukemogenic NUP98-HOXA9 and SET-NUP214 Fusion Proteins Using a Proteomic Approach.
Mendes A, Jühlen R, Bousbata S, Fahrenkrog B. Mendes A, et al. Cells. 2020 Jul 10;9(7):1666. doi: 10.3390/cells9071666. Cells. 2020. PMID: 32664447 Free PMC article. - Evolution of a transcriptional regulator from a transmembrane nucleoporin.
Franks TM, Benner C, Narvaiza I, Marchetto MC, Young JM, Malik HS, Gage FH, Hetzer MW. Franks TM, et al. Genes Dev. 2016 May 15;30(10):1155-71. doi: 10.1101/gad.280941.116. Epub 2016 May 19. Genes Dev. 2016. PMID: 27198230 Free PMC article. - The Oncogenic Fusion Proteins SET-Nup214 and Sequestosome-1 (SQSTM1)-Nup214 Form Dynamic Nuclear Bodies and Differentially Affect Nuclear Protein and Poly(A)+ RNA Export.
Port SA, Mendes A, Valkova C, Spillner C, Fahrenkrog B, Kaether C, Kehlenbach RH. Port SA, et al. J Biol Chem. 2016 Oct 28;291(44):23068-23083. doi: 10.1074/jbc.M116.735340. Epub 2016 Sep 9. J Biol Chem. 2016. PMID: 27613868 Free PMC article. - Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1.
Crampton N, Kodiha M, Shrivastava S, Umar R, Stochaj U. Crampton N, et al. Mol Biol Cell. 2009 Dec;20(24):5106-16. doi: 10.1091/mbc.e09-05-0397. Mol Biol Cell. 2009. PMID: 19828735 Free PMC article.
References
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113:1651–1659. - PubMed
- Azuma Y, Dasso M. The role of Ran in nuclear function. Curr Opin Cell Biol. 2000;12:302–307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases