Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage - PubMed (original) (raw)
Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage
Jeroen Essers et al. EMBO J. 2002.
Abstract
Recombination between homologous DNA molecules is essential for the proper maintenance and duplication of the genome, and for the repair of exogenously induced DNA damage such as double-strand breaks. Homologous recombination requires the RAD52 group proteins, including Rad51, Rad52 and Rad54. Upon treatment of mammalian cells with ionizing radiation, these proteins accumulate into foci at sites of DNA damage induction. We show that these foci are dynamic structures of which Rad51 is a stably associated core component, whereas Rad52 and Rad54 rapidly and reversibly interact with the structure. Furthermore, we show that the majority of the proteins are not part of the same multi-protein complex in the absence of DNA damage. Executing DNA transactions through dynamic multi-protein complexes, rather than stable holo-complexes, allows flexibility. In the case of DNA repair, for example, it will facilitate cross-talk between different DNA repair pathways and coupling to other DNA transactions, such as replication.
Figures
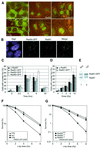
Fig. 1. DNA damage response of the human RAD52 group proteins in living cells. (A) Detection of Rad51–GFP, Rad52–GFP and Rad54–GFP in living CHO cells before treatment with ionizing radiation by a combination of fluorescence and phase-contrast microscopy (0 Gy; upper panels). All three proteins formed nuclear foci upon treatment with ionizing radiation (12 Gy; lower panels). Images were taken 2 h after irradiation. (B) DNA damage-induced colocalization of Rad51 and Rad54–GFP. Cells expressing Rad54–GFP were fixed 2 h after treatment with ionizing radiation (12 Gy). Nuclei were visualized by DAPI staining. The signal from Rad54–GFP (shown in green) was observed directly by fluorescence microscopy. Endogenous Rad51 (shown in red) was detected by indirect immunofluorescence using antibodies against Rad51. Colocalization of Rad51 and Rad54–GFP (shown in yellow) is evident in the merged image. (C) Kinetics of endogenous Rad51, Rad51–GFP, Rad52–GFP and Rad54–GFP DNA damage-induced foci formation. CHO cells or their derivatives expressing the indicated GFP-tagged RAD52 group proteins were fixed at the indicated times after treatment with ionizing radiation (12 Gy). Detection of foci was performed as described in (B). The percentage of cells containing nuclear foci of endogenous Rad51, Rad51–GFP, Rad52–GFP or Rad54–GFP was determined in three independent experiments. (D) Dose response of endogenous and Rad51–GFP foci. CHO cells and their derivative expressing Rad51–GFP were fixed 2 h after treatment with the indicated doses of ionizing radiation. Detection of foci was performed as described above. (E) Immunoblot of endogenous and Rad51–GFP. Cell-free extracts prepared from V79 cells stably expressing Rad51–GFP were fractionated into nuclear (Nuc) and cytoplasmic (Cyt) fractions, which were analyzed for the presence of endogenous Rad51 and Rad51–GFP by immunoblotting using antibodies against Rad51. (F) Clonogenic survival assays of V79 cells and their indicated derivatives. V79 cells and V79 cells expressing Rad51–GFP were equally sensitive to ionizing radiation as measured by colony-forming ability after irradiation. A derivative of V79 (irs1), defective in the Rad51 paralogue Xrcc2, served as a control for irradiation. (G) Clonogenic survival assays of the Rad54-proficient and -deficient cells after ionizing radiation. The ionizing radiation sensitivity of _Rad54_–/– mouse embryonic stem cells was corrected to wild-type levels by the expression of Rad54–GFP.
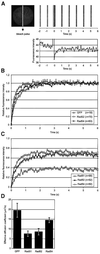
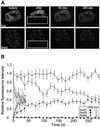
Fig. 2. Fluorescence redistribution after photobleaching (FRAP) analyses of RAD52 group proteins in CHO cells. Cells stably expressing Rad51–GFP, Rad52–GFP and Rad54–GFP were subjected to a local bleach pulse, and the kinetics of fluorescence recovery in the bleached area was determined. (A) An example of the primary data obtained using the photobleaching protocol on a cell nucleus (shown left) containing Rad54–GFP. The fluorescence in a small strip (indicated by the hatched rectangle) spanning the entire nucleus was bleached with a 200 ms high-intensity laser pulse. The recovery of fluorescence in the strip was monitored at intervals of 100 ms. For clarity, only strips obtained every second are shown above the time scale of the experiment. The measured fluorescence intensities over time are plotted below. (B) The photobleaching protocol was applied to a number (n) of cells containing GFP, Rad52–GFP and Rad54–GFP. The fluorescence intensity immediately after bleaching was set to 0, and the final post-bleach pulse fluorescence intensity measured was set to 1. The normalized data are plotted. (C) The photobleaching protocol was applied to a number (n) of cells containing Rad51–GFP, Rad52–GFP and Rad54–GFP. In this case, the final measured fluorescence intensity was normalized to the pre-bleach pulse fluorescence intensity. (D) The effective diffusion coefficients of the RAD52 group proteins were determined by fitting the experimentally obtained curves [shown in (B) and (C)] to a mathematical model describing diffusion (see Materials and methods). Measurements were performed in triplicate, and consistent results were obtained among different sets of experiments. Error bars indicate twice the standard error of the mean.
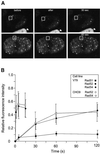
Fig. 3. Different residence times of RAD52 group proteins in DNA damage-induced foci. (A) Individual DNA damage-induced foci (indicated by squares) in cells stably expressing Rad52–GFP were photobleached. Images were collected before, immediately after and at the indicated times after the bleach pulse. (B) Quantitative FRAP analysis of DNA damage-induced Rad51–GFP, Rad52–GFP and Rad54–GFP foci. Recovery of fluorescence was measured at the indicated time points after the bleach pulse. All data points represent the mean of at least 10 different measurements, and the error bars indicate twice the standard error of the mean. The results were independent of the cell line used, because similar results were obtained with V79 and CHO9 cells. The major cause of fluctuations in fluorescence intensity of the foci was cellular movement.
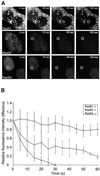
Fig. 4. FLIP in DNA damage-induced Rad51–GFP, Rad52–GFP and Rad54–GFP foci. (A) A region (indicated by circles) in the nucleoplasm of ionizing radiation-treated (12 Gy) cells expressing Rad51–GFP, Rad52–GFP or Rad54–GFP was repeatedly bleached 2 h after the irradiation. Cells were imaged between bleach pulses at the indicated times after the initial bleach pulse. (B) Quantitative FLIP analysis of DNA damage-induced foci containing Rad51–GFP, Rad52–GFP and Rad54–GFP. The difference between the loss in fluorescence of the DNA damage-induced foci and that of the nucleoplasm was determined at the indicated time points after the initial bleach pulse. The resulting curves were corrected for background bleaching due to monitoring of the cells. For each data point, at least five different cells and five foci per cell were analyzed. Error bars indicate twice the standard error of the mean.
Fig. 5. FRAP and FLIP in DNA damage-induced Rad51–GFP, Rad52–GFP and Rad54–GFP foci. (A) A region (indicated by rectangles) of a cell containing Rad52–GFP foci was bleached by a single laser pulse (upper panels). The cell was imaged at the indicated times after bleaching. FLIP was measured in foci in the unbleached half of the cell, and FRAP was measured in foci in the bleached half of the same cell. The same experimental protocol applied to a fixed cell demonstrates the requirement for protein mobility to observe FRAP and FLIP (lower panels). (B) Quantitation of the simultaneous FRAP/FLIP experiment on DNA damage-induced Rad51–GFP, Rad52–GFP and Rad54–GFP foci. Error bars indicate twice the standard error of the mean.
Similar articles
- Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52.
van Veelen LR, Essers J, van de Rakt MW, Odijk H, Pastink A, Zdzienicka MZ, Paulusma CC, Kanaar R. van Veelen LR, et al. Mutat Res. 2005 Jul 1;574(1-2):34-49. doi: 10.1016/j.mrfmmm.2005.01.020. Epub 2005 Apr 9. Mutat Res. 2005. PMID: 15914205 - Analysis of DNA recombination and repair proteins in living cells by photobleaching microscopy.
Essers J, Houtsmuller AB, Kanaar R. Essers J, et al. Methods Enzymol. 2006;408:463-85. doi: 10.1016/S0076-6879(06)08029-3. Methods Enzymol. 2006. PMID: 16793387 - Xrcc3 is recruited to DNA double strand breaks early and independent of Rad51.
Forget AL, Bennett BT, Knight KL. Forget AL, et al. J Cell Biochem. 2004 Oct 15;93(3):429-36. doi: 10.1002/jcb.20232. J Cell Biochem. 2004. PMID: 15372620 - [DNA homologous recombination repair in mammalian cells].
Popławski T, Błasiak J. Popławski T, et al. Postepy Biochem. 2006;52(2):180-93. Postepy Biochem. 2006. PMID: 17078508 Review. Polish. - [Analyses of recA Rad51-like gene deficient mice].
Yoshida K, Morita T. Yoshida K, et al. Tanpakushitsu Kakusan Koso. 2001 Jun;46(8 Suppl):1021-9. Tanpakushitsu Kakusan Koso. 2001. PMID: 11436289 Review. Japanese. No abstract available.
Cited by
- The Use of Survival Dose-Rate Dependencies as Theoretical Discrimination Criteria for In-Silico Dynamic Radiobiological Models.
Mingo Barba S, Lobo-Cerna F, Krawczyk PM, Lattuada M, Füchslin RM, Petri-Fink A, Scheidegger S. Mingo Barba S, et al. Dose Response. 2024 Aug 30;22(3):15593258241279906. doi: 10.1177/15593258241279906. eCollection 2024 Jul-Sep. Dose Response. 2024. PMID: 39224699 Free PMC article. - Distinct mobility patterns of BRCA2 molecules at DNA damage sites.
Paul MW, Aaron J, Wait E, Van Genderen RM, Tyagi A, Kabbech H, Smal I, Chew TL, Kanaar R, Wyman C. Paul MW, et al. Nucleic Acids Res. 2024 Aug 12;52(14):8332-8343. doi: 10.1093/nar/gkae559. Nucleic Acids Res. 2024. PMID: 38953170 Free PMC article. - Liquid-liquid phase separation in DNA double-strand breaks repair.
Wang YL, Zhao WW, Shi J, Wan XB, Zheng J, Fan XJ. Wang YL, et al. Cell Death Dis. 2023 Nov 15;14(11):746. doi: 10.1038/s41419-023-06267-0. Cell Death Dis. 2023. PMID: 37968256 Free PMC article. Review. - In vivo tracking of functionally tagged Rad51 unveils a robust strategy of homology search.
Liu S, Miné-Hattab J, Villemeur M, Guerois R, Pinholt HD, Mirny LA, Taddei A. Liu S, et al. Nat Struct Mol Biol. 2023 Oct;30(10):1582-1591. doi: 10.1038/s41594-023-01065-w. Epub 2023 Aug 21. Nat Struct Mol Biol. 2023. PMID: 37605042 - Repair Foci as Liquid Phase Separation: Evidence and Limitations.
Miné-Hattab J, Liu S, Taddei A. Miné-Hattab J, et al. Genes (Basel). 2022 Oct 13;13(10):1846. doi: 10.3390/genes13101846. Genes (Basel). 2022. PMID: 36292731 Free PMC article. Review.
References
- Baumann P. and West,S.C. (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci., 23, 247–251. - PubMed
- Bishop D.K., Ear,U., Bhattacharyya,A., Calderone,C., Beckett,M., Weichselbaum,R.R. and Shinohara,A. (1998) Xrcc3 is required for assembly of Rad51 complexes in vivo. J. Biol. Chem., 273, 21482–21488. - PubMed
- Chen G. et al. (1999) Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J. Biol. Chem., 274, 12748–12752. - PubMed
- Cremer T. and Cremer,C. (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Rev. Genet., 2, 292–301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials