Isoform-specific interaction of HP1 with human TAFII130 - PubMed (original) (raw)
Isoform-specific interaction of HP1 with human TAFII130
Milo F Vassallo et al. Proc Natl Acad Sci U S A. 2002.
Abstract
The general transcription factor TFIID facilitates recruitment of the transcription machinery to gene promoters and regulates initiation of transcription by RNA polymerase II. hTAF(II)130, a component of TFIID, interacts with and serves as a coactivator for multiple transcriptional regulatory proteins, including Sp1 and CREB. A yeast two-hybrid screen has identified an interaction between hTAF(II)130 and heterochromatin protein 1 (HP1), a chromatin-associated protein whose function has been implicated in gene silencing. We find that hTAF(II)130 associates with HP1 in an isoform-specific manner: HP1alpha and HP1gamma bind to hTAF(II)130, but not HP1beta. In addition, we show that endogenous hTAF(II)130 and components of TFIID in HeLa nuclear extracts associate with glutathione S-transferase-HP1alpha and -HP1gamma. hTAF(II)130 possesses a pentapeptide HP1-binding motif, and mutation of the hTAF(II)130 HP1 box compromises the interaction of hTAF(II)130 with HP1. We demonstrate that Gal4-HP1 proteins interfere with hTAF(II)130-mediated activation of transcription. Our results suggest that HP1alpha and HP1gamma associate with hTAF(II)130 to mediate repression of transcription, supporting a new model of transcriptional repression involving a specific interaction between a component of TFIID and chromatin.
Figures
Figure 1
Multiple clones of human HP1α and HP1γ are isolated in a yeast two-hybrid screen by using the central domain of hTAFII130 as bait. (A) A schematic representation of the bait protein LexA-hTAFII130N/C composed of the DNA-binding domain (DBD) of LexA fused to residues 270–700 of hTAFII130 (numbering according to ref. 7). Conserved region I (CI), conserved region II (CII), and glutamine-rich regions (Q1 to Q4) are indicated. (B) A schematic representation of HP1α and HP1γ clones isolated from the yeast screen. Arrows demarcate the sequence boundaries for each clone. The 3′ end of all isolated HP1 clones included the complete C-terminal coding sequence of HP1. The clone numbers and their amino acid positions are indicated for each isolate. The smallest clone contained only the chromoshadow domain of HP1γ. Protein interaction was not detectable between HP1 and LexA DBD alone or with other transcription factors. The expression of HA-tagged HP1 proteins was confirmed in yeast cell lysates by immunoblotting with α-HA antibody.
Figure 2
The HP1 C-terminal domain is required for its association with hTAFII130 in vitro. (A) A schematic representation of the GST-HP1 fusion proteins used in the study. (B) _In vitro_-translated hTAFII 130N/C was incubated with comparable amounts of the indicated GST fusion proteins. The result (summarized in A at right) suggests that an intact chromoshadow domain of HP1γ is required for hTAFII130 interaction. GST-BE is a control fusion protein containing a small fragment of hTAFII130 similar in size to HP1γ.
Figure 3
hTAFII130 binds to HP1 through an HP1 box present in hTAFII130. (A) hTAFII130 derivatives were translated in vitro and incubated with GST-HP1γ. Relative amounts of hTAFII130 derivative bound to GST-HP1γ are indicated qualitatively at right. The thick vertical bar represents newly identified HP1 interaction motif (HP1 box) present in all hTAFII130 constructs that associate with HP1. (B) An alignment of HP1-associated protein sequences containing an HP1 box (43). Conserved residues (consensus PXVXL where X is any amino acid) are boxed. *, residues that, when mutated, compromised the binding of each protein to HP1 in vitro. hTAFII130-DE is a mutant with alterations of the two conserved amino acids in the HP1 box. (C) Point mutations in the HP1 box compromised hTAFII130 binding to HP1γ. _In vitro_-translated wild type and the DE mutant of hTAFII130N/C was incubated with GST-HP1γ. A comparable amount of GST-BE was used as a control for nonspecific binding. (D) hTAFII130 bound to HP1α and HP1γ but not to HP1β. GST fusions of HP1α, HP1β, and HP1γ were incubated with _in vitro_-translated hTAFII130 (1–947). The bound fractions were separated by SDS/PAGE and visualized by autoradiography.
Figure 4
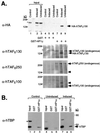
Endogenous TAFIIs and TBP from HeLa cells bind to GST-HP1. (A) Nuclear extracts were prepared from control HeLa cells (lanes 1, 4, and 7), and a HeLa cell line induced (lanes 2, 5, and 8) or uninduced (lanes 3, 6, and 9) for HA-hTAFII130 expression. The extracts were incubated with GST (lanes 4–6) or GST-HP1γ (lanes 7–9); bound proteins were analyzed by sequential immunoblotting with α-HA, α-hTAFII130, α-hTAFII250, and α-hTAFII100 antibody. Endogenous hTAFII130 is larger in size than the recombinant HA-hTAFII130, which lacks the extreme N-terminal sequence. The recombinant HA-hTAFII130 in the induced cell extract is detected in the fraction bound to HP1γ, whereas endogenous hTAFII130, hTAFII250, and hTAFII100 were detected in all three nuclear extracts bound to HP1γ. The arrowheads in the third and fourth panels correspond to HA-hTAFII130, whose signal remained after sequential immunoblotting with the monoclonal antibodies. *, nonspecific background bands. (B) The same nuclear extracts used in A were incubated with GST or GST-HP1α, and the bound fractions were analyzed for the presence of hTBP with α-hTBP antibody.
Figure 5
HP1 interferes with hTAFII130-mediated stimulation of transcription. (A) HeLa cells transfected with a plasmid-expressing hTAFII130 stimulated UAS-Luc reporter containing two Gal4-binding sites upstream of the minimal angiotensinogen promoter (lane 2). This activation was inhibited by the coexpression of Gal4-HP1γ (lane 3) but not by Gal4-HP1γΔC (lane 4). An unrelated repressor protein Gal4-LANA also did not block activation by hTAFII130 (lane 5). (B) HeLa cells were cotransfected with 2XGal/2XLex-E1bTATA-luciferase reporter plasmid and plasmids expressing LexA-hTAFII130N/C and/or Gal4-HP1, as indicated. Gal4-HP1α and Gal4-HP1γ more potently inhibited LexA-hTAFII130N/C-mediated activation compared with Gal4-HP1β.
Similar articles
- Genetic dissection of hTAF(II)130 defines a hydrophobic surface required for interaction with glutamine-rich activators.
Rojo-Niersbach E, Furukawa T, Tanese N. Rojo-Niersbach E, et al. J Biol Chem. 1999 Nov 19;274(47):33778-84. doi: 10.1074/jbc.274.47.33778. J Biol Chem. 1999. PMID: 10559271 - Sp1 and AP2 regulate but do not constitute TATA-less human TAF(II)55 core promoter activity.
Zhou T, Chiang CM. Zhou T, et al. Nucleic Acids Res. 2002 Oct 1;30(19):4145-57. doi: 10.1093/nar/gkf537. Nucleic Acids Res. 2002. PMID: 12364593 Free PMC article. - The coactivator dTAF(II)110/hTAF(II)135 is sufficient to recruit a polymerase complex and activate basal transcription mediated by CREB.
Felinski EA, Quinn PG. Felinski EA, et al. Proc Natl Acad Sci U S A. 2001 Nov 6;98(23):13078-83. doi: 10.1073/pnas.241337698. Epub 2001 Oct 30. Proc Natl Acad Sci U S A. 2001. PMID: 11687654 Free PMC article. - Function of heterochromatin protein 1 during DNA repair.
Bártová E, Malyšková B, Komůrková D, Legartová S, Suchánková J, Krejčí J, Kozubek S. Bártová E, et al. Protoplasma. 2017 May;254(3):1233-1240. doi: 10.1007/s00709-017-1090-3. Epub 2017 Feb 24. Protoplasma. 2017. PMID: 28236007 Review. - Mechanisms of functional promiscuity by HP1 proteins.
Canzio D, Larson A, Narlikar GJ. Canzio D, et al. Trends Cell Biol. 2014 Jun;24(6):377-86. doi: 10.1016/j.tcb.2014.01.002. Epub 2014 Mar 4. Trends Cell Biol. 2014. PMID: 24618358 Free PMC article. Review.
Cited by
- The Heterochromatin Protein 1 family.
Lomberk G, Wallrath L, Urrutia R. Lomberk G, et al. Genome Biol. 2006;7(7):228. doi: 10.1186/gb-2006-7-7-228. Genome Biol. 2006. PMID: 17224041 Free PMC article. Review. - Diversity in TAF proteomics: consequences for cellular differentiation and migration.
Kazantseva J, Palm K. Kazantseva J, et al. Int J Mol Sci. 2014 Sep 19;15(9):16680-97. doi: 10.3390/ijms150916680. Int J Mol Sci. 2014. PMID: 25244017 Free PMC article. Review. - Polycomb repressive complex 2 and H3K27me3 cooperate with H3K9 methylation to maintain heterochromatin protein 1α at chromatin.
Boros J, Arnoult N, Stroobant V, Collet JF, Decottignies A. Boros J, et al. Mol Cell Biol. 2014 Oct 1;34(19):3662-74. doi: 10.1128/MCB.00205-14. Epub 2014 Jul 21. Mol Cell Biol. 2014. PMID: 25047840 Free PMC article. - BCL11A-dependent recruitment of SIRT1 to a promoter template in mammalian cells results in histone deacetylation and transcriptional repression.
Senawong T, Peterson VJ, Leid M. Senawong T, et al. Arch Biochem Biophys. 2005 Feb 15;434(2):316-25. doi: 10.1016/j.abb.2004.10.028. Arch Biochem Biophys. 2005. PMID: 15639232 Free PMC article. - TAF10 Interacts with the GATA1 Transcription Factor and Controls Mouse Erythropoiesis.
Papadopoulos P, Gutiérrez L, Demmers J, Scheer E, Pourfarzad F, Papageorgiou DN, Karkoulia E, Strouboulis J, van de Werken HJ, van der Linden R, Vandenberghe P, Dekkers DH, Philipsen S, Grosveld F, Tora L. Papadopoulos P, et al. Mol Cell Biol. 2015 Jun;35(12):2103-18. doi: 10.1128/MCB.01370-14. Epub 2015 Apr 13. Mol Cell Biol. 2015. PMID: 25870109 Free PMC article.
References
- Albright S R, Tjian R. Gene. 2000;242:1–13. - PubMed
- Bell B, Tora L. Exp Cell Res. 1999;246:11–19. - PubMed
- Struhl K, Moqtaderi Z. Cell. 1998;94:1–4. - PubMed
- Green M R. Trends Biochem Sci. 2000;25:59–63. - PubMed
- Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Cell. 1993;72:247–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials