In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex - PubMed (original) (raw)
In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex
Arina D Omer et al. Proc Natl Acad Sci U S A. 2002.
Abstract
The genomes of hyperthermophilic Archaea encode dozens of methylation guide, C/D box small RNAs that guide 2'-O-methylation of ribose to specific sites in rRNA and various tRNAs. The genes encoding the Sulfolobus homologues of eukaryotic proteins that are known to be present in C/D box small nucleolar ribonucleoprotein (snoRNP) complexes were cloned, and the proteins (aFIB, aNOP56, and aL7a) were expressed and purified. The purified proteins along with an in vitro transcript of the Sulfolobus sR1 small RNA were reconstituted in vitro, into an RNP complex. The order of assembly of the three proteins onto the RNA was aL7a, aNOP56, and aFIB. The complex was active in targeting S-adenosyl methionine (SAM)-dependent, site-specific 2'-O-methylation of ribose to a short fragment of ribosomal RNA (rRNA) that was complementary to the D box guide region of the sR1 small RNA. The presence of aFIB was essential for methylation; mutant proteins having amino acid replacements in the SAM-binding motif of aFIB were able to assemble into an RNP complex, but the resulting complexes were defective in methylation activity. These experiments define the minimal number of components and the conditions required to achieve in vitro RNA guide-directed 2'-O-methylation of ribose in a target RNA.
Figures
Figure 1
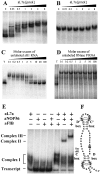
In vitro assembly of archaeal sR1 sRNA into an RNP complex. In vitro transcribed RNAs were uniformly labeled with [α-32P]ATP and used in gel mobility retardation assays to monitor interaction with recombinant aL7a protein. S. acidocaldarius C/D box sR1 sRNA (0.2 pmol) (A), or RNase P RNA (1 pmol) (B), was mixed with increasing amounts of recombinant aL7a protein (0 to 8 pmol per assay) at 0°C, transferred to 70°C for 10 min, separated on a nondenaturing 10% polyacrylamide gel, and visualized by autoradiography. The competition assays contained radiolabeled sR1 RNA (0.2 pmol), aL7a protein (1 pmol), and nonradiolabeled competitor sR1 (C), or RNase P (D) RNAs (0.02 to 20 pmol). T, transcript. To detect higher-order complexes, uniformly labeled sR1 transcript (0.2 pmol) was mixed with one or more of the proteins (1 pmol of each protein per reaction) at 0°C, transferred to 70°C for 10 min, separated on a nondenaturing 6% polyacrylamide gel, and visualized by autoradiography (E). The positions of free transcript, complex I (sR1 sRNA–aL7a), complex II (sR1 sRNA–aL7a–aNOP56), and complex III (sR1 sRNA–aL7a–aNOP56–aFIB) are indicated. A secondary structural model of sR1s RNA is depicted (F). The aL7a protein is predicted to bind to the loops generated by the C/D or C′/D′ motifs (indicated by *). The base predicted to rotate out of the loop and insert into the pocket of the protein is the first U residue in the C or C′ box sequence and is highlighted in black (20).
Figure 2
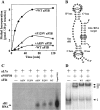
RNP guide-dependent methyl incorporation into a complementary target RNA. RNP complex was assembled by renaturing _in vitro_transcribed sR1 sRNA and target RNA (5′-GGGAUAAGCCA[U]GGGAGUCUUACACUCCC-3′; the expected site for methylation is bracketed) and mixing with aFIB, aNOP56, aL7a, and [_methyl_-3H]SAM at 0°C. The mixture (120 μl containing 720 pmol of sR1 guide RNA, 720 pmol of target RNA, 360 pmol of radioactive SAM, and 24 pmol of each of the three recombinant proteins) was divided into six 20-μl reactions and transferred to 70°C. At time intervals, single 20-μl reactions were removed and precipitated at 0°C with 5% trichloroacetic acid. The precipitates were collected on nitrocellulose filters, and dried, and radioactivity was determined by scintillation counting. The kinetics of methyl incorporation are shown on the left (A) and the predicted secondary structure of the guide and target RNAs are shown on the right (B). The wild type (wt) and the two mutant aFIB proteins (containing A85V or P129V replacements) were tested for their ability to assemble into RNP particles by using 3′-end-labeled sR1 sRNA transcript (1 pmol) and coimmunoprecipitation with antibodies against aFIB. The coprecipitated RNAs were displayed on a denaturing 6% polyacrylamide gel and detected by autoradiography (C). The ability of the A85V mutant to assemble into complex III (D) was examined as described in the legend to Fig. 1.
Figure 3
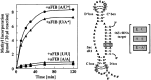
Effects of nucleotide substitution in the methylation guide and target RNA sequences. Nucleotide substitutions were introduced into the sR1 D box guide (A to U at the N plus five position relative to the start of the D box) and the rRNA target (U to A at position corresponding to U52 in 16S rRNA). Methylation assays, as described in the legend to Fig. 2, were carried out by using (i) wild-type guide and wild-type target (●), (ii) mutant guide and wild-type target (♦), (iii) wild-type guide and mutant target (■), or (iv) compensatory mutant guide and mutant target (▴). The base pair at the methylation site for each assay is illustrated on the right and the incorporation kinetics for the four reactions is illustrated on the left.
Figure 4
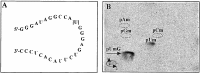
Thin-layer chromatographic separation of the hydrolysis products of the target RNA. The standard 29-nt long target RNA contains a single unique UG dinucleotide. The U residue in this dinucleotide is the expected site of methylation and corresponds to position U52 in 16S rRNA (A). Guide and target RNAs were mixed in a 20-μl reaction in the standard methylation assay using [_methyl_-3H]SAM and incubated at 70°C for 1 h. The RNA was extracted, digested with 0.01 unit of P1 nuclease (Roche Diagnostics) for 12 h at 37°C, and products were mixed with 5 nmol each of pAm, pCm, pGm, pUm, and pUmG standards before the two-dimensional TLC separation (28). Unlabeled standards were detected by UV shading and radioactivity was detected by 3H-imaging (B). The spots corresponding to the UV-detectable standards were excised from the plate and subjected to scintillation counting (see text). Omission of any of the RNA or protein components from the reaction mixture results in background levels of incorporation of radioactivity into acid-insoluble material (see Fig. 2).
Similar articles
- RNA-guided nucleotide modification of ribosomal and non-ribosomal RNAs in Archaea.
Ziesche SM, Omer AD, Dennis PP. Ziesche SM, et al. Mol Microbiol. 2004 Nov;54(4):980-93. doi: 10.1111/j.1365-2958.2004.04319.x. Mol Microbiol. 2004. PMID: 15522081 - Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes.
Gaspin C, Cavaillé J, Erauso G, Bachellerie JP. Gaspin C, et al. J Mol Biol. 2000 Apr 7;297(4):895-906. doi: 10.1006/jmbi.2000.3593. J Mol Biol. 2000. PMID: 10736225 - C/D box sRNA-guided 2'-O-methylation patterns of archaeal rRNA molecules.
Dennis PP, Tripp V, Lui L, Lowe T, Randau L. Dennis PP, et al. BMC Genomics. 2015 Aug 22;16:632. doi: 10.1186/s12864-015-1839-z. BMC Genomics. 2015. PMID: 26296872 Free PMC article. - Small non-coding RNAs in Archaea.
Dennis PP, Omer A. Dennis PP, et al. Curr Opin Microbiol. 2005 Dec;8(6):685-94. doi: 10.1016/j.mib.2005.10.013. Epub 2005 Oct 26. Curr Opin Microbiol. 2005. PMID: 16256421 Review. - The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA.
Watkins NJ, Bohnsack MT. Watkins NJ, et al. Wiley Interdiscip Rev RNA. 2012 May-Jun;3(3):397-414. doi: 10.1002/wrna.117. Epub 2011 Nov 7. Wiley Interdiscip Rev RNA. 2012. PMID: 22065625 Review.
Cited by
- Insights into the mechanism of ribosomal incorporation of mammalian L13a protein during ribosome biogenesis.
Das P, Basu A, Biswas A, Poddar D, Andrews J, Barik S, Komar AA, Mazumder B. Das P, et al. Mol Cell Biol. 2013 Aug;33(15):2829-42. doi: 10.1128/MCB.00250-13. Epub 2013 May 20. Mol Cell Biol. 2013. PMID: 23689135 Free PMC article. - The structure and function of small nucleolar ribonucleoproteins.
Reichow SL, Hamma T, Ferré-D'Amaré AR, Varani G. Reichow SL, et al. Nucleic Acids Res. 2007;35(5):1452-64. doi: 10.1093/nar/gkl1172. Epub 2007 Feb 6. Nucleic Acids Res. 2007. PMID: 17284456 Free PMC article. Review. - Structural organization of box C/D RNA-guided RNA methyltransferase.
Ye K, Jia R, Lin J, Ju M, Peng J, Xu A, Zhang L. Ye K, et al. Proc Natl Acad Sci U S A. 2009 Aug 18;106(33):13808-13. doi: 10.1073/pnas.0905128106. Epub 2009 Aug 5. Proc Natl Acad Sci U S A. 2009. PMID: 19666563 Free PMC article. - Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation.
Charpentier B, Muller S, Branlant C. Charpentier B, et al. Nucleic Acids Res. 2005 Jun 2;33(10):3133-44. doi: 10.1093/nar/gki630. Print 2005. Nucleic Acids Res. 2005. PMID: 15933208 Free PMC article. - The coiled-coil domain of the Nop56/58 core protein is dispensable for sRNP assembly but is critical for archaeal box C/D sRNP-guided nucleotide methylation.
Zhang X, Champion EA, Tran EJ, Brown BA 2nd, Baserga SJ, Maxwell ES. Zhang X, et al. RNA. 2006 Jun;12(6):1092-103. doi: 10.1261/rna.2230106. Epub 2006 Apr 6. RNA. 2006. PMID: 16601205 Free PMC article.
References
- Maxwell E S, Fournier M J. Annu Rev Biochem. 1995;64:897–934. - PubMed
- Lafontaine D L J, Tollervey D. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 281–288.
- Bachellerie J-P, Cavaille J, Qu L-H. In: The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. Garrett R A, Douthwaite S R, Liljas A, Matheson A T, Moore P B, Noller H F, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 199–203.
- Venema J, Tollervey D. Annu Rev Genet. 1999;33:261–311. - PubMed
- Filipowicz W, Pelczar P, Pogacic V, Dragon F. Acta Biochim Pol. 1999;46:377–389. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources