Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity - PubMed (original) (raw)
Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity
Francesco Parlati et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Syntaxin-5 (Sed5) is the only syntaxin needed for transport into and across the yeast Golgi, raising the question of how a single syntaxin species could mediate vesicle transport in both the anterograde and the retrograde direction within the stack. Sed5 is known to combine with two light chains (Bos1 and Sec22) to form the t-SNARE needed to receive vesicles from the endoplasmic reticulum. However, the yeast Golgi contains several other potential light chains with which Sed5 could potentially combine to form other t-SNAREs. To explore the degree of specificity in the choice of light chains by a t-SNARE, we undertook a comprehensive examination of the capacity of all 21 Sed5-based t-SNAREs that theoretically could assemble in the yeast Golgi to fuse with each of the 7 potential v-SNAREs also present in this organelle. Only one additional of these 147 combinations was fusogenic. This functional proteomic strategy thereby revealed a previously uncharacterized t-SNARE in which Sed5 is the heavy chain and Gos1 and Ykt6 are the light chains, and whose unique cognate v-SNARE is Sft1. Immunoprecipitation experiments confirmed the existence of this complex in vivo. Fusion mediated by this second Golgi SNAREpin is topologically restricted, and existing genetic and morphologic evidence implies that it is used for transport across the Golgi stack. From this study, together with the previous functional proteomic analyses which have tested 275 distinct quaternary SNARE combinations, it follows that the fusion potential and transport pathways of the yeast cell can be read out from its genome sequence according to the SNARE hypothesis with a predictive accuracy of about 99.6%.
Figures
Figure 1
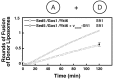
Two tetramers of Golgi-localized SNARE proteins catalyze membrane fusion. (A) Twenty-one combinations of 3 Golgi-localized SNARE proteins were reconstituted into liposomes and analyzed by SDS/PAGE (Lanes 1–21). All combinations contain the Golgi heavy chain Sed5. (B) Golgi-localized SNAREs Bet1, Bos1, Sec22, Sft1, Gos1, Vti1, and Ykt6 (Lanes 1–7, respectively) were individually reconstituted into donor liposomes; reconstituted protein was analyzed by SDS/PAGE. (C) Membrane-fusion assay identifies two fusogenic sets of donor and acceptor liposomes of 147 possible combinations. Twenty-one different acceptor liposomes (described in upper right box) were mixed with either Bet1, Bos1, Sec22, Vti1, Ykt6, Gos1, or Sft-containing donor liposomes, and the increase in fluorescence was monitored over two h. The increase in fluorescence was converted into a measure of rounds of fusion of donor liposomes.
Figure 2
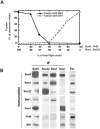
(A) Sed5, Gos1, Ykt6, and Sft1 are necessary and sufficient to mediate lipid bilayer fusion. Sed5, Gos1, Ykt6 (Lane 1), Sed5, Gos1 (Lane 2), Sed5, Ykt6 (Lane 3), and Gos1, Ykt6 (Lane 4) were reconstituted into acceptor liposomes and analyzed by SDS/PAGE. These acceptor liposomes were mixed with Sft1 donor liposomes at 37°C, and the increase in fluorescence was monitored and converted to rounds of fusion (top graph). Fusion between acceptor and donor liposomes is v-SNARE- and t-SNARE-dependent and can be blocked by the soluble domain of Sft1 (bottom graph). Acceptor liposomes containing either Sed5/Gos1,Ykt6 or no protein (p.f.) were mixed with donor liposomes containing either Sft1 or no protein (p.f.) in the presence of either Sft1 cytosolic domain (Sft1c) or buffer (buf). (B) Topological-restriction of fusion mediated by t-Sed5/Gos1,Ykt6 and v-Sft1. A 4:0 topology (Bottom) and a 2:2 topology (Middle) of Golgi SNAREs do not catalyze membrane fusion. A distinct 3:1 topology (Top) of Golgi SNAREs mediates membrane fusion. Golgi SNAREs were reconstituted into acceptor liposomes or donor liposomes, mixed, and the increase in fluorescence was monitored over 2 h at 37°C.
Figure 3
Specificity of fusion between the Golgi t-SNARE (Sed5/Gos1,Ykt6) and v-SNARE (Sft1) liposomes. Eleven potential v-SNAREs encoded in the yeast genome were independently reconstituted into donor liposomes and mixed with Sed5/Gos1,Ykt6 acceptor liposomes. A background of 0.072 rounds of fusion, representing fusion with protein-free liposomes, was subtracted from each experiment. The extent of fusion was subsequently normalized to represent 100% fusion for the cognate v-SNARE Sft1.
Figure 4
A peptide corresponding to the C-terminal domain of Sft1 stimulates fusion. Soluble peptide corresponding to the C-terminal portion of the Sft1 SNARE domain, vCOOH-Sft1 (see_Materials and Methods_) can stimulate fusion between Sed5/Gos1,Ykt6 acceptor liposomes and Sft1 donor liposomes. As a control, vCOOH-peptides corresponding to Bet1, Bos1, Sec22, Ykt6, Gos1, Vti1, Nyv1, Tlg1, and Snc2 were tested, and the 120-min time point was averaged (black square). Sed5/Gos1,Ykt6 acceptor liposomes were mixed with Sft1 donor liposomes in the absence or presence of Sft1 peptide. Increase in fluorescence was monitored at 37°C over 2 h and converted into a measure of rounds of fusion.
Figure 5
(A) Shifting Sed5 between its two t-SNAREs according to the prevailing composition of light chains. Acceptor liposomes containing a constant amount of Sed5, Gos1, and Ykt6 with increasing amounts of Bos1 and Sec22 were reconstituted and mixed with Sft1 donor liposomes (○). Conversely, acceptor liposomes containing Sed5, Bos1, and Sec22 with increasing amounts of Gos1 and Ykt6 were reconstituted and mixed with Bet1 donor liposomes (●). The increase in fluorescence was monitored and converted to a measure of rounds of fusion. (B) Sed5 forms two distinct Golgi SNARE complexes in vivo. Sec18–1 spheroplasts were preincubated for 1 h at the restrictive temperature of 37°C and then lysed in detergent in the presence of 2 mM EDTA (see Materials and Methods). Cell extracts (10 mg of total protein) were incubated with antibodies to indicated SNAREs (IP) covalently coupled to protein A agarose; the resulting immunoprecipitates were washed with lysis buffer containing 0.3 M KCl. Proteins were eluted with 0.2 M glycine pH 2.7, precipitated with trichloroacetic acid, resolved by SDS/PAGE, and then subjected to an immunoblot analysis using antibodies directed against the indicated SNAREs (Immunoblot). Immunoprecipitated material (10%) and 0.1% of total cell extract (Ext) were loaded onto the gel.
Similar articles
- Topological restriction of SNARE-dependent membrane fusion.
Parlati F, McNew JA, Fukuda R, Miller R, Söllner TH, Rothman JE. Parlati F, et al. Nature. 2000 Sep 14;407(6801):194-8. doi: 10.1038/35025076. Nature. 2000. PMID: 11001058 - Compartmental specificity of cellular membrane fusion encoded in SNARE proteins.
McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Söllner TH, Rothman JE. McNew JA, et al. Nature. 2000 Sep 14;407(6801):153-9. doi: 10.1038/35025000. Nature. 2000. PMID: 11001046 - A t-SNARE of the endocytic pathway must be activated for fusion.
Paumet F, Brügger B, Parlati F, McNew JA, Söllner TH, Rothman JE. Paumet F, et al. J Cell Biol. 2001 Dec 10;155(6):961-8. doi: 10.1083/jcb.200104092. Epub 2001 Dec 10. J Cell Biol. 2001. PMID: 11739407 Free PMC article. - SNAREs and membrane fusion in the Golgi apparatus.
Nichols BJ, Pelham HR. Nichols BJ, et al. Biochim Biophys Acta. 1998 Aug 14;1404(1-2):9-31. doi: 10.1016/s0167-4889(98)00044-5. Biochim Biophys Acta. 1998. PMID: 9714710 Review. - Fusion of membranes during the acrosome reaction: a tale of two SNAREs.
Kierszenbaum AL. Kierszenbaum AL. Mol Reprod Dev. 2000 Dec;57(4):309-10. doi: 10.1002/1098-2795(200012)57:4<309::AID-MRD1>3.0.CO;2-W. Mol Reprod Dev. 2000. PMID: 11066058 Review.
Cited by
- SNARE-Mediated Exocytosis in Neuronal Development.
Urbina FL, Gupton SL. Urbina FL, et al. Front Mol Neurosci. 2020 Aug 7;13:133. doi: 10.3389/fnmol.2020.00133. eCollection 2020. Front Mol Neurosci. 2020. PMID: 32848598 Free PMC article. Review. - The SNARE Protein Syntaxin 3 Confers Specificity for Polarized Axonal Trafficking in Neurons.
Soo Hoo L, Banna CD, Radeke CM, Sharma N, Albertolle ME, Low SH, Weimbs T, Vandenberg CA. Soo Hoo L, et al. PLoS One. 2016 Sep 23;11(9):e0163671. doi: 10.1371/journal.pone.0163671. eCollection 2016. PLoS One. 2016. PMID: 27662481 Free PMC article. - COPI activity coupled with fatty acid biosynthesis is required for viral replication.
Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. Cherry S, et al. PLoS Pathog. 2006 Oct;2(10):e102. doi: 10.1371/journal.ppat.0020102. PLoS Pathog. 2006. PMID: 17040126 Free PMC article. - A SNARE required for retrograde transport to the endoplasmic reticulum.
Burri L, Varlamov O, Doege CA, Hofmann K, Beilharz T, Rothman JE, Söllner TH, Lithgow T. Burri L, et al. Proc Natl Acad Sci U S A. 2003 Aug 19;100(17):9873-7. doi: 10.1073/pnas.1734000100. Epub 2003 Jul 31. Proc Natl Acad Sci U S A. 2003. PMID: 12893879 Free PMC article. - Sugary Logistics Gone Wrong: Membrane Trafficking and Congenital Disorders of Glycosylation.
Linders PTA, Peters E, Ter Beest M, Lefeber DJ, van den Bogaart G. Linders PTA, et al. Int J Mol Sci. 2020 Jun 30;21(13):4654. doi: 10.3390/ijms21134654. Int J Mol Sci. 2020. PMID: 32629928 Free PMC article. Review.
References
- Söllner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. Nature (London) 1993;362:318–324. - PubMed
- Weber T, Zemelman B V, McNew J A, Westermann B, Gmachl M, Parlati F, Söllner T H, Rothman J E. Cell. 1998;92:759–772. - PubMed
- Pelham H R. Trends Cell Biol. 2001;11:99–101. - PubMed
- Chen Y A, Scheller R H. Nat Rev Mol Cell Biol. 2001;2:98–106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases