Identification of genes expressed with temporal-spatial restriction to developing cerebellar neuron precursors by a functional genomic approach - PubMed (original) (raw)
Identification of genes expressed with temporal-spatial restriction to developing cerebellar neuron precursors by a functional genomic approach
Qing Zhao et al. Proc Natl Acad Sci U S A. 2002.
Erratum in
- Proc Natl Acad Sci U S A 2002 Jun 11;99(12):8460
Abstract
Hedgehog pathway activation is required for proliferation of cerebellar granule cell neuron precursors during development and is etiologic in certain cerebellar tumors. To identify genes expressed specifically in granule cell neuron precursors, we used oligonucleotide microarrays to analyze regulation of 13,179 genes/expressed sequence tags in heterogeneous primary cultures of neonatal mouse cerebellum that respond to the mitogen Sonic hedgehog. In conjunction, we applied experiment-specific noise models to render a gene-by-gene robust indication of up-regulation in Sonic hedgehog-treated cultures. Twelve genes so identified were tested, and 10 (83%) showed appropriate expression in the external granular layer (EGL) of the postnatal day (PN) 7 cerebellum and down-regulation by PN 15, as verified by in situ hybridization. Whole-organ profiling of the developing cerebellum was carried out from PN 1 to 30 to generate a database of temporal gene regulation profiles (TRPs). From the database an algorithm was developed to capture the TRP typical of EGL-specific genes. The "TRP-EGL" accurately predicted expression in vivo of an additional 18 genes/expressed sequence tags with a sensitivity of 80% and a specificity of 88%. We then compared the positive predictive value of our analytical procedure with other widely used methods, as verified by the TRP-EGL in silico. These findings suggest that replicate experiments and incorporation of noise models increase analytical specificity. They further show that genome-wide methods are an effective means to identify stage-specific gene expression in the developing granule cell lineage.
Figures
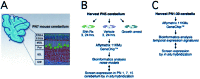
Figure 1
Structure of PN 7 mouse cerebellum and experimental approach. (A) The layered architecture of the cerebellum in a parasagittal section. The EGL comprises actively proliferating granule cells (EGLa) and those that recently have left the cell cycle (EGLb). The Purkinje cell (Pur), molecular (MOL), internal granule cell (IGL), and cerebellar white matter (CW) layers are indicated. (B) Schematic diagram of experimental strategy used to identify 76 genes/EST significantly up-regulated by Shh under various culture conditions and verification by ISH. (C) Whole cerebella during PN1–30 were analyzed by GeneChips to generate a database of temporal gene regulation used to validate results of analytical techniques in silico.
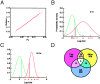
Figure 2
Genes up-regulated under experimental conditions. (A) Distribution of measurement error. The y axis is the output of Gaussian random number generator N (0, 0.0065), and the x axis is μN from 5,000 permutations τ of condition labels. The linear relationship between_X_ and Y indicates Gaussian distribution of measurement error. (B and C) Example of the log-fold distribution caused by noise (FDN; green line) and log-fold distribution of Shh-treated vs. vehicle-treated cells (red line) at 3 (B) and 24 (C) h for the gene_cdc20_. Areas underneath the curves indicate respective probabilities. μ, mean; σ, SD. (D) Total number of genes/EST significantly up-regulated at 3 (red) and 24 (yellow) h and under conditions of growth arrest (blue).
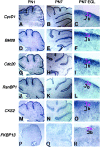
Figure 3
Expression of candidate genes/ESTs in vivo in the neonatal mouse cerebellum. Representative results of ISH showing the gene-expression pattern of identified genes in the PN 1 and 7 postnatal mouse cerebellum at low power magnification (×40) are shown. (C, F, I,L, O, and R) The right-hand column shows high-power magnification (×400) images of the EGL and delineates the actively proliferating granule cells (from those that recently have left the cell cycle) (Fig. 1_A_). The gene-expression pattern for the group I genes cycD1 (A_–_C), BM28 (D_–_F), cdc20(G_–_I), RanBP1(J_–_L), and CKS2(M_–_O) indicated mRNA transcripts in the EGLa. In contrast, genes such as FKBP13(P_–_R) did not show expression in PN 7 EGL.
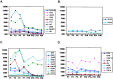
Figure 4
Temporal analysis of candidate genes/EST expression by whole-organ expression profiling of mouse neonatal cerebellum. (A) Graphical representation of temporal regulation of the 10 of 12 genes from the training set with confirmed expression in the cerebellar EGL (Table 1). Note that they show a similar pattern comprising relatively high levels of expression at PN 3–7 relative to PN 15–30. (B) Temporal analysis of the remaining 2 of 12 genes (Table 1) from the training set. (C and_D_) Pattern of temporal regulation of 18 additional genes tested (“test set,” Table 1). Note that 8 of 10 genes (C) displayed a similar pattern to genes in_A_. Conversely, HMG1 exhibited a late peak at PN 15. Seven of nine genes (D) showed a nonspecific temporal pattern of regulation. It should be noted that the failure to identify expression of EST69 and _FKBP13_could be because of technical reasons such as faulty probe design or low endogenous levels of expression in vivo.x axis, duplicate samples of mouse cerebella harvested at PNs 1, 3, 5, 7, 10, 15, 21, and 30 were analyzed; _y_axis, absolute value of Avg Diff as reported by Affymetrix software (scale set at 2,000).
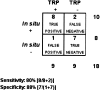
Figure 5
Sensitivity and specificity of using temporal gene-expression pattern_in silico_ to predict expression of candidate genes/EST in vivo. As shown, the TRP results in an overall sensitivity of 80% and specificity of 88%. Assuming the stochastic independence of the in situ confirmation outcome and the temporal gene regulatory pattern, a χ2 test of the contingency table gives a probability (P < 0.01) that the association between confirmation and pattern is insignificant.
Similar articles
- Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum.
Wallace VA. Wallace VA. Curr Biol. 1999 Apr 22;9(8):445-8. doi: 10.1016/s0960-9822(99)80195-x. Curr Biol. 1999. PMID: 10226030 - Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling.
Rios I, Alvarez-Rodríguez R, Martí E, Pons S. Rios I, et al. Development. 2004 Jul;131(13):3159-68. doi: 10.1242/dev.01188. Development. 2004. PMID: 15197161 - Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors.
Kenney AM, Cole MD, Rowitch DH. Kenney AM, et al. Development. 2003 Jan;130(1):15-28. doi: 10.1242/dev.00182. Development. 2003. PMID: 12441288 - Pituitary adenylate cyclase-activating polypeptide and sonic hedgehog interact to control cerebellar granule precursor cell proliferation.
Nicot A, Lelièvre V, Tam J, Waschek JA, DiCicco-Bloom E. Nicot A, et al. J Neurosci. 2002 Nov 1;22(21):9244-54. doi: 10.1523/JNEUROSCI.22-21-09244.2002. J Neurosci. 2002. PMID: 12417650 Free PMC article. - Growth arrest specific gene 1: a fuel for driving growth in the cerebellum.
Marques G, Fan CM. Marques G, et al. Cerebellum. 2002 Dec;1(4):259-63. doi: 10.1080/147342202320883560. Cerebellum. 2002. PMID: 12879964 Review.
Cited by
- The Gs-linked receptor GPR3 inhibits the proliferation of cerebellar granule cells during postnatal development.
Tanaka S, Shaikh IM, Chiocca EA, Saeki Y. Tanaka S, et al. PLoS One. 2009 Jun 15;4(6):e5922. doi: 10.1371/journal.pone.0005922. PLoS One. 2009. PMID: 19526062 Free PMC article. - An essential role for p38 MAPK in cerebellar granule neuron precursor proliferation.
Guldal CG, Ahmad A, Korshunov A, Squatrito M, Awan A, Mainwaring LA, Bhatia B, Parathath SR, Nahle Z, Pfister S, Kenney AM. Guldal CG, et al. Acta Neuropathol. 2012 Apr;123(4):573-86. doi: 10.1007/s00401-012-0946-z. Acta Neuropathol. 2012. PMID: 22302101 Free PMC article. - Gene expression profiling of Duchenne muscular dystrophy skeletal muscle.
Haslett JN, Sanoudou D, Kho AT, Han M, Bennett RR, Kohane IS, Beggs AH, Kunkel LM. Haslett JN, et al. Neurogenetics. 2003 Aug;4(4):163-71. doi: 10.1007/s10048-003-0148-x. Epub 2003 Apr 16. Neurogenetics. 2003. PMID: 12698323 - Chemokine receptor CXCR4 as a therapeutic target for neuroectodermal tumors.
Shim H, Oishi S, Fujii N. Shim H, et al. Semin Cancer Biol. 2009 Apr;19(2):123-34. doi: 10.1016/j.semcancer.2008.11.004. Epub 2008 Nov 25. Semin Cancer Biol. 2009. PMID: 19084067 Free PMC article. Review. - SHH pathway and cerebellar development.
Vaillant C, Monard D. Vaillant C, et al. Cerebellum. 2009 Sep;8(3):291-301. doi: 10.1007/s12311-009-0094-8. Epub 2009 Feb 18. Cerebellum. 2009. PMID: 19224309 Review.
References
- Edlund T, Jessell T M. Cell. 1999;96:211–224. - PubMed
- Geschwind D H, Ou J, Easterday M C, Dougherty J D, Jackson R L, Chen Z, Antoine H, Terskikh A, Weissman I L, Nelson S F, Kornblum H I. Neuron. 2001;29:325–339. - PubMed
- Dent G W, O'Dell D M, Eberwine J H. Physiol Behav. 2001;73:841–847. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases