Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription - PubMed (original) (raw)
Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription
Douglas Kyung Nam et al. Proc Natl Acad Sci U S A. 2002.
Abstract
We have analyzed a systematic flaw in the current system of gene identification: the oligo(dT) primer widely used for cDNA synthesis generates a high frequency of truncated cDNAs through internal poly(A) priming. Such truncated cDNAs may contribute to 12% of the expressed sequence tags in the current dbEST database. By using a synthetic transcript and real mRNA templates as models, we characterized the patterns of internal poly(A) priming by oligo(dT) primer. We further demonstrated that the internal poly(A) priming can be effectively diminished by replacing the oligo(dT) primer with a set of anchored oligo(dT) primers for reverse transcription. Our study indicates that cDNAs designed for genomewide gene identification should be synthesized by use of the anchored oligo(dT) primers, rather than the oligo(dT) primers, to diminish the generation of truncated cDNAs caused by internal poly(A) priming.
Figures
Figure 1
Schematics of internal poly(A) priming by oligo(dT) primer. Besides priming the poly(A) sequence at the 3′ end of mRNA, the oligo(dT) primer can also prime the poly(A) sequences present internally within mRNA. The initiation of cDNA synthesis from the oligo(dT) primed at both locations results in the generation of two truncated cDNAs from a single mRNA template.
Figure 2
Comparison of internal poly(A) priming and 3′ poly(A) priming between oligo(dT) primer and anchored oligo(dT) primers. An in vitro transcript containing an internal A20 and 3′ A50 was used for reverse transcription with either oligo(dT) primer or anchored oligo(dT) primers. (A) Sequencing image of cDNA clones primed by oligo(dT) primer and anchored oligo(dT) primers. (B) Summary of the results of the analyses.
Figure 3
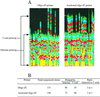
Internal poly(A) priming prevents the extension of 3′ end-primed cDNA along the mRNA template. A group of mRNAs with internal poly(A) sequences was primed with oligo(dT) or anchored oligo(dT) primers for reverse transcription. Quantitative PCR was used to determine cDNA levels originating from internal poly(A) priming, and 3′ end poly(A) priming extending beyond the internal poly(A) tract. (A) Demonstration of the levels of cDNA from internal poly(A) priming and 3′ end poly(A) priming in proteoglycan mRNA. It shows the diminishing of cDNA originating from 3′ end priming when oligo(dT) primer was used, whereas the level was high between internal and 3′ end-primed cDNA when anchored oligo(dT) primers were used. (B) Summary of the results from the analyses. The value from internal priming is set as 1 for comparing the relative levels between the cDNA from internal priming and cDNA from external priming.
Figure 4
Effects of high-dose oligo(dT) primers on the internal poly(A) priming and 3′ end poly(A) priming. mRNA from proteoglycan with 10 internal As was used in this analysis. Increased doses of oligo(dT) primer were used for the first-strand cDNA synthesis. Quantitative PCR was used to determine cDNA levels originating from internal poly(A) priming and 3′ end poly(A) priming extending beyond the internal poly(A) tract. The same amount of control templates was used in the quantitative PCR for each set of reactions.
Figure 5
Estimated prevalence of ESTs in dbEST originating from internal poly(A) priming. (A) Summary of the analyses. See the text for detailed description. (B) Distance between the sequence matched by EST to the internal poly(A) sequence in the matched reference sequence.
Similar articles
- cDNA amplification using one-sided (anchored) PCR.
Dorit RL, Ohara O. Dorit RL, et al. Curr Protoc Immunol. 2001 May;Chapter 10:Unit 10.24. doi: 10.1002/0471142735.im1024s08. Curr Protoc Immunol. 2001. PMID: 18432689 - Priming reverse transcription with oligo(dT) does not yield representative samples of Mycobacterium tuberculosis cDNA.
Lakey DL, Zhang Y, Talaat AM, Samten B, DesJardin LE, Eisenach KD, Johnston SA, Barnes PF. Lakey DL, et al. Microbiology (Reading). 2002 Aug;148(Pt 8):2567-2572. doi: 10.1099/00221287-148-8-2567. Microbiology (Reading). 2002. PMID: 12177350 - Evidence for polyadenylated mRNA in Pseudomonas aeruginosa.
Saravanamuthu SS, von Götz F, Salunkhe P, Chozhavendan R, Geffers R, Buer J, Tümmler B, Steinmetz I. Saravanamuthu SS, et al. J Bacteriol. 2004 Oct;186(20):7015-8. doi: 10.1128/JB.186.20.7015-7018.2004. J Bacteriol. 2004. PMID: 15466054 Free PMC article. - Isolation of Poly(A)+ Messenger RNA Using Magnetic Oligo(dT) Beads.
Green MR, Sambrook J. Green MR, et al. Cold Spring Harb Protoc. 2019 Oct 1;2019(10). doi: 10.1101/pdb.prot101733. Cold Spring Harb Protoc. 2019. PMID: 31575797 - Polyadenylylation in mycobacteria: evidence for oligo(dT)-primed cDNA synthesis.
Adilakshmi T, Ayling PD, Ratledge C. Adilakshmi T, et al. Microbiology (Reading). 2000 Mar;146 ( Pt 3):633-638. doi: 10.1099/00221287-146-3-633. Microbiology (Reading). 2000. PMID: 10746766
Cited by
- Effects of doxycycline on gene expression in Wolbachia and Brugia malayi adult female worms in vivo.
Rao RU, Huang Y, Abubucker S, Heinz M, Crosby SD, Mitreva M, Weil GJ. Rao RU, et al. J Biomed Sci. 2012 Feb 9;19(1):21. doi: 10.1186/1423-0127-19-21. J Biomed Sci. 2012. PMID: 22321609 Free PMC article. - A survey of transcriptome complexity using full-length isoform sequencing in the tea plant Camellia sinensis.
Ma D, Fang J, Ding Q, Wei L, Li Y, Zhang L, Zhang X. Ma D, et al. Mol Genet Genomics. 2022 Sep;297(5):1243-1255. doi: 10.1007/s00438-022-01913-2. Epub 2022 Jun 28. Mol Genet Genomics. 2022. PMID: 35763065 - Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq.
Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Shepard PJ, et al. RNA. 2011 Apr;17(4):761-72. doi: 10.1261/rna.2581711. Epub 2011 Feb 22. RNA. 2011. PMID: 21343387 Free PMC article. - Squaramides and Ureas: A Flexible Approach to Polymerase-Compatible Nucleic Acid Assembly.
Shivalingam A, Taemaitree L, El-Sagheer AH, Brown T. Shivalingam A, et al. Angew Chem Int Ed Engl. 2020 Jul 6;59(28):11416-11422. doi: 10.1002/anie.202000209. Epub 2020 May 7. Angew Chem Int Ed Engl. 2020. PMID: 32153132 Free PMC article. - Comparative analysis of alternative polyadenylation in S. cerevisiae and S. pombe.
Liu X, Hoque M, Larochelle M, Lemay JF, Yurko N, Manley JL, Bachand F, Tian B. Liu X, et al. Genome Res. 2017 Oct;27(10):1685-1695. doi: 10.1101/gr.222331.117. Epub 2017 Sep 15. Genome Res. 2017. PMID: 28916539 Free PMC article.
References
- International Human Genome Sequencing Consortium. Nature (London) 2001;409:860–921. - PubMed
- Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. Nature (London) 2001;409:685–690. - PubMed
- Adams M D, Kerlavage A R, Fields C, Venter J C. Nat Genet. 1993;4:256–267. - PubMed
- Hillier L D, Lennon G, Becker M, Bonaldo M F, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Genome Res. 1996;6:807–828. - PubMed
- Aaronson J S, Eckman B, Blevins R A, Borkowski J A, Myerson J, Imran S, Elliston K O. Genome Res. 1996;6:829–845. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources