Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome - PubMed (original) (raw)
Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome
Peter Kosarev et al. Genome Biol. 2002.
Abstract
Background: In computational analysis, the RING-finger domain is one of the most frequently detected domains in the Arabidopsis proteome. In fact, it is more abundant in Arabidopsis than in other eukaryotic genomes. However, computational analysis might classify ambiguous domains of the closely related PHD and LIM motifs as RING domains by mistake. Thus, we set out to define an ordered set of Arabidopsis RING domains by evaluating predicted domains on the basis of recent structural data.
Results: Inspection of the proteome with a current InterPro release predicts 446 RING domains. We evaluated each detected domain and as a result eliminated 59 false positives. The remaining 387 domains were grouped by cluster analysis and according to their metal-ligand arrangement. We further defined novel patterns for additional computational analyses of the proteome. They were based on recent structural data that enable discrimination between the related RING, PHD and LIM domains. These patterns allow us to predict with different degrees of certainty whether a particular domain is indeed likely to form a RING finger.
Conclusions: In summary, 387 domains have a significant potential to form a RING-type cross-brace structure. Many of these RING domains overlap with predicted PHD domains; however, the RING domain signature mostly prevails. Thus, the abundance of PHD domains in Arabidopsis has been significantly overestimated. Cluster analysis of the RING domains defines groups of proteins, which frequently show significant similarity outside the RING domain. These groups document a common evolutionary origin of their members and potentially represent genes of overlapping functionality.
Figures
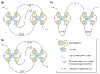
Figure 1
Schematic presentation of the structure of prototypical RING, PHD and LIM domains. The metal-ligand residues, either cysteine (C) or histidine (H), are shown as numbered spheres. Two pairs of metal ligands coordinate one zinc ion (hexagon). The numbers next to the loops connecting the metal-ligand residues indicate the minimum and maximum number of loop residues. (a) The structure of a RING domain (RING-HC type). The metal-ligand pairs 1 and 3 coordinate one zinc ion, while pairs 2 and 4 coordinate the second one in a so-called cross-brace arrangement. (b) The structure of a PHD domain reveals a cross-brace arrangement similar to the RING domain. (c) The LIM domain structure is distinct in its consecutive zinc ligation scheme: the first zinc ion is coordinated by the metal-ligand pairs 1 and 2, while the second ion is coordinated by pairs 3 and 4.
Similar articles
- Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome.
Englbrecht CC, Schoof H, Böhm S. Englbrecht CC, et al. BMC Genomics. 2004 Jul 5;5(1):39. doi: 10.1186/1471-2164-5-39. BMC Genomics. 2004. PMID: 15236668 Free PMC article. - Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis.
Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. Stone SL, et al. Plant Physiol. 2005 Jan;137(1):13-30. doi: 10.1104/pp.104.052423. Plant Physiol. 2005. PMID: 15644464 Free PMC article. - [Comparative phylogenetic analysis of the rice and Arabidopsis PHD-finger proteins].
Feng Y, Liu QP, Xue QZ. Feng Y, et al. Yi Chuan Xue Bao. 2004 Nov;31(11):1284-93. Yi Chuan Xue Bao. 2004. PMID: 15651682 Chinese. - PDZ domain proteins: 'dark matter' of the plant proteome?
Gardiner J, Overall R, Marc J. Gardiner J, et al. Mol Plant. 2011 Nov;4(6):933-7. doi: 10.1093/mp/ssr043. Epub 2011 Jun 7. Mol Plant. 2011. PMID: 21653283 Review. - ATLs and BTLs, plant-specific and general eukaryotic structurally-related E3 ubiquitin ligases.
Guzmán P. Guzmán P. Plant Sci. 2014 Feb;215-216:69-75. doi: 10.1016/j.plantsci.2013.10.017. Epub 2013 Nov 5. Plant Sci. 2014. PMID: 24388516 Review.
Cited by
- Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa).
Chai G, Hu R, Zhang D, Qi G, Zuo R, Cao Y, Chen P, Kong Y, Zhou G. Chai G, et al. BMC Genomics. 2012 Jun 18;13:253. doi: 10.1186/1471-2164-13-253. BMC Genomics. 2012. PMID: 22708723 Free PMC article. - Characterization and promoter analysis of a cotton RING-type ubiquitin ligase (E3) gene.
Ho MH, Saha S, Jenkins JN, Ma DP. Ho MH, et al. Mol Biotechnol. 2010 Oct;46(2):140-8. doi: 10.1007/s12033-010-9280-7. Mol Biotechnol. 2010. PMID: 20376576 - A gene family encoding RING finger proteins in rice: their expansion, expression diversity, and co-expressed genes.
Lim SD, Yim WC, Moon JC, Kim DS, Lee BM, Jang CS. Lim SD, et al. Plant Mol Biol. 2010 Mar;72(4-5):369-80. doi: 10.1007/s11103-009-9576-9. Epub 2009 Dec 3. Plant Mol Biol. 2010. PMID: 19957018 - The CCCH-Type Zinc-Finger Protein GhC3H20 Enhances Salt Stress Tolerance in Arabidopsis thaliana and Cotton through ABA Signal Transduction Pathway.
Zhang Q, Zhang J, Wei F, Fu X, Wei H, Lu J, Ma L, Wang H. Zhang Q, et al. Int J Mol Sci. 2023 Mar 6;24(5):5057. doi: 10.3390/ijms24055057. Int J Mol Sci. 2023. PMID: 36902489 Free PMC article. - Zooming into the structure-function of RING finger proteins for anti-cancer therapeutic applications.
George M, Masamba P, Iwalokun BA, Kappo AP. George M, et al. Am J Cancer Res. 2023 Jul 15;13(7):2773-2789. eCollection 2023. Am J Cancer Res. 2023. PMID: 37559981 Free PMC article. Review.
References
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. InterPro - an integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16:1145–1150. - PubMed
- Freemont PS, Hanson IM, Trowsdale J. A novel cysteine-rich sequence motif. Cell. 1991;64:483–484. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases