Abnormal development of the hippocampal dentate gyrus in mice lacking the CXCR4 chemokine receptor - PubMed (original) (raw)
Abnormal development of the hippocampal dentate gyrus in mice lacking the CXCR4 chemokine receptor
Meiling Lu et al. Proc Natl Acad Sci U S A. 2002.
Abstract
We investigated the role of the CXCR4 chemokine receptor in development of the mouse hippocampus. CXCR4 mRNA is expressed at sites of neuronal and progenitor cell migration in the hippocampus at late embryonic and early postnatal ages. mRNA for stromal cell-derived factor 1 (SDF-1), the only known ligand for the CXCR4 receptor, is expressed close to these migration sites, in the meninges investing the hippocampal primordium and the primordium itself. In mice engineered to lack the CXCR4 receptor, the morphology of the hippocampal dentate gyrus (DG) is dramatically altered. Gene expression markers for DG granule neurons and bromodeoxyuridine labeling of dividing cells revealed an underlying defect in the stream of postmitotic cells and secondary dentate progenitor cells that migrate toward and form the DG. In the absence of CXCR4, the number of dividing cells in the migratory stream and in the DG itself is reduced, and neurons appear to differentiate prematurely before reaching their target. Our findings indicate a role for the SDF-1/CXCR4 chemokine signaling system in DG morphogenesis. Finally, the DG is unusual as a site of adult neurogenesis. We find that both CXCR4 and SDF-1 are expressed in the adult DG, suggesting an ongoing role in DG morphogenesis.
Figures
Figure 1
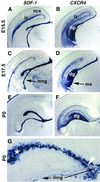
Expression of SDF-1 and CXCR4 in developing neocortex and hippocampus. (A_–_G) Coronal sections through mouse forebrains on E15.5, E17.5, and the day of birth (P0) processed for in situ hybridization. G is a higher magnification of the DG (dg) region. SDF-1 is expressed in the intermediate zone (iz) of the neocortex (ncx) (A), in the meninges (mng) overlying the hippocampus (hp) (A, C, E, and G), and in a zone close to the hippocampal fissure also occupied by Cajal–Retzius cells (A, C, E, and G; see text). White arrowhead in G points to the opening of the hippocampal fissure. White asterisk in G marks some of the _SDF-1_-expressing cells lining the hippocampal fissure. CXCR4 also is expressed in the iz at E15.5 (B). At all ages, strongest CXCR4 expression appears in the developing dg and in a migratory stream (ms) of cells moving toward the dg (D and F).
Figure 2
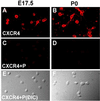
Acutely isolated hippocampal neurons express CXCR4 receptors. (A_–_D) Confocal microscopy images of E17.5 and P0 hippocampal neurons immunostained for CXCR4 antibody. (A and B) Anti-CXCR4 antibody staining. Immunostaining was diminished after preadsorption of the CXCR4 antibody with its peptide antigen (C and D). (E and F) Differential interference contrast microscopy images of the same field of cells as C and D.
Figure 3
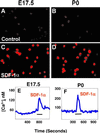
Effect of SDF-1α on CREB activation and calcium mobilization in acutely isolated E17.5 and P0 hippocampal neurons. (A_–_D) Fluorescence microscopy image of neurons immunoreactive for the activated form of CREB. Neurons were treated with PBS (control) or SDF-1α for 10 min in Eagle's minimal essential medium. (A and B) No CREB activation in neurons treated with PBS. (C and D) Activation of CREB in neurons treated with 100 nM SDF-1α. (E and F) SDF-1α increase [Ca2+]i in hippocampal neurons. SDF-1α was applied for the times indicated by the bars (3–4 min). Representative traces from single neurons are shown in E and F. On E17.5, 8 of 127 hippocampal neurons were responsive to SDF-1α. By P0, 11 of 194 neurons were responsive to SDF-1α.
Figure 4
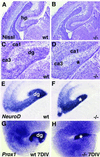
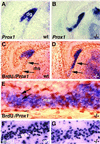
Abnormal development of the DG in mice deficient in CXCR4. (A_–_D) Nissl-stained coronal sections through E18.5 wild-type (A and C) or homozygote CXCR4 mutant forebrains (B and D). C and D are higher-magnification views of A and B. Gross morphology appears similar between wild-type and mutant brains in the developing neocortex (A and B) and the developing CA fields (ca1, ca3) of the hippocampal pyramidal cell layer (C and D). By contrast, the developing dg is distinct in the wild type (C) but almost undetectable in the mutant (asterisk in D). (E and F) Coronal sections processed with in situ hybridization to show expression of NeuroD. The developing dg is distinct in a wild-type mouse (E) but not in a homozygote CXCR4 mutant (asterisk in F). (G and H) Hippocampal slices prepared from an E18.5 wild-type (G) or CXCR4 mutant mouse (H) and maintained for 7 days in vitro (7DIV). Expression of Prox1 reveals a classic, V-shaped dg in the wild-type slice. By contrast, the CXCR4 mutant slice shows a small, poorly formed dg (asterisk in H).
Figure 5
Defects in the secondary proliferative cell population that forms the DG. (A_–_D) Coronal sections through the E18.5 hippocampus of wild-type (A and C) or CXCR4 mutant mice (B and D), processed to show expression of Prox1 (blue) or Prox1 together with BrdUrd-labeled dividing cells (orange). In the wild-type mouse, Prox1 is expressed in the forming dg (A and C). By contrast, in the mutant, Prox1 is expressed in the vestigial dg but also along the migratory stream (ms) (arrows in D) of dividing cells running along the ventral surface of the hippocampus into the dg. BrdUrd-labeled cells of the ms can be seen between the two arrows in C. (E) A higher magnification of the migrating stream of cells shown in D. Numerous blue, _Prox1_-expressing cells appear among the brown, BrdUrd-labeled cells, but the populations appear largely distinct. Arrows in E point to two single-labeled cells. (F and G) High-magnification views of BrdUrd-labeled dividing cells (dark blue) coursing through the ms in a wild-type (F) and a CXCR4 mutant mouse (G). About 30% fewer BrdUrd-labeled cells appear in the ms of the mutant (G) than in the wild type (F).
Figure 6
Expression of SDF-1 and CXCR4 in the adult mouse DG. (A_–_C) Coronal sections through the hippocampus of CD-1 adult mice. CXCR4 is expressed in scattered cells in the dg hilus and lining the dg blades (arrows point to four such cells) (A). Double labeling with BrdUrd (brown) suggests that some of the _CXCR4_-expressing (blue) cells are still dividing (B). Arrowheads in B indicate double-labeled cells; singly BrdUrd-labeled or _CXCR4-_expressing cells appear in the same field. SDF-1 is expressed in the blades of the dg, and scattered SDF-1-expressing cells (arrows) also appear in the hilus of the dg (C).
Similar articles
- Expression of SDF-1 and CXCR4 during reorganization of the postnatal dentate gyrus.
Berger O, Li G, Han SM, Paredes M, Pleasure SJ. Berger O, et al. Dev Neurosci. 2007;29(1-2):48-58. doi: 10.1159/000096210. Dev Neurosci. 2007. PMID: 17148948 - The chemokine stromal cell-derived factor-1 regulates GABAergic inputs to neural progenitors in the postnatal dentate gyrus.
Bhattacharyya BJ, Banisadr G, Jung H, Ren D, Cronshaw DG, Zou Y, Miller RJ. Bhattacharyya BJ, et al. J Neurosci. 2008 Jun 25;28(26):6720-30. doi: 10.1523/JNEUROSCI.1677-08.2008. J Neurosci. 2008. PMID: 18579746 Free PMC article. - COUP-TFI mitotically regulates production and migration of dentate granule cells and modulates hippocampal Cxcr4 expression.
Parisot J, Flore G, Bertacchi M, Studer M. Parisot J, et al. Development. 2017 Jun 1;144(11):2045-2058. doi: 10.1242/dev.139949. Epub 2017 May 15. Development. 2017. PMID: 28506990 - Morphogenesis of the dentate gyrus: what we are learning from mouse mutants.
Li G, Pleasure SJ. Li G, et al. Dev Neurosci. 2005 Mar-Aug;27(2-4):93-9. doi: 10.1159/000085980. Dev Neurosci. 2005. PMID: 16046842 Review.
Cited by
- The immune system and developmental programming of brain and behavior.
Bilbo SD, Schwarz JM. Bilbo SD, et al. Front Neuroendocrinol. 2012 Aug;33(3):267-86. doi: 10.1016/j.yfrne.2012.08.006. Epub 2012 Sep 9. Front Neuroendocrinol. 2012. PMID: 22982535 Free PMC article. Review. - Signals Orchestrating Peripheral Nerve Repair.
Rigoni M, Negro S. Rigoni M, et al. Cells. 2020 Jul 24;9(8):1768. doi: 10.3390/cells9081768. Cells. 2020. PMID: 32722089 Free PMC article. Review. - Axonal Regeneration after Spinal Cord Injury: Molecular Mechanisms, Regulatory Pathways, and Novel Strategies.
Elmalky MI, Alvarez-Bolado G, Younsi A, Skutella T. Elmalky MI, et al. Biology (Basel). 2024 Sep 7;13(9):703. doi: 10.3390/biology13090703. Biology (Basel). 2024. PMID: 39336130 Free PMC article. Review. - Dentate gyrus morphogenesis is regulated by β-catenin function in hem-derived fimbrial glia.
Parichha A, Datta D, Suresh V, Chatterjee M, Holtzman MJ, Tole S. Parichha A, et al. Development. 2022 Nov 1;149(21):dev200953. doi: 10.1242/dev.200953. Epub 2022 Oct 21. Development. 2022. PMID: 36196585 Free PMC article. - Stromal derived factor-1 exerts differential regulation on distinct cortical cell populations in vitro.
Pritchett J, Wright C, Zeef L, Nadarajah B. Pritchett J, et al. BMC Dev Biol. 2007 Apr 10;7:31. doi: 10.1186/1471-213X-7-31. BMC Dev Biol. 2007. PMID: 17425785 Free PMC article.
References
- Rossi D, Zlotnik A. Annu Rev Immunol. 2000;18:217–242. - PubMed
- Berger E A, Murphy P M, Farber J M. Annu Rev Immunol. 1999;17:657–700. - PubMed
- Bajetto A, Bonavia R, Barbero S, Florio T, Schettini G. Front Neuroendocrinol. 2001;22:147–184. - PubMed
- Miller R, Oh S. In: Universes in Delicate Balance; Chemokines in the Nervous System. Ransohoff R, editor. Amsterdam: Elsevier Science; 2002. pp. 273–288.
- Zou Y R, Kottmann A H, Kuroda M, Taniuchi I, Littman D R. Nature (London) 1998;393:595–599. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MH40165/MH/NIMH NIH HHS/United States
- DA13141/DA/NIDA NIH HHS/United States
- R37 MH059962/MH/NIMH NIH HHS/United States
- R01 MH059962/MH/NIMH NIH HHS/United States
- DK44840/DK/NIDDK NIH HHS/United States
- R01 DA013141/DA/NIDA NIH HHS/United States
- R37 MH040165/MH/NIMH NIH HHS/United States
- MH59962/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases