Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE - PubMed (original) (raw)
Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE
Zonglin Hu et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Selection of the division site in Escherichia coli is regulated by the min system and requires the rapid oscillation of MinD between the two halves of the cell under the control of MinE. In this study we have further investigated the molecular basis for this oscillation by examining the interaction of MinD with phospholipid vesicles. We found that MinD bound to phospholipid vesicles in the presence of ATP and, upon binding, assembled into a well-ordered helical array that deformed the vesicles into tubes. Stimulation of the MinD ATPase by addition of MinE led to disassembly of the tubes and the release of MinD from the vesicles. It is proposed that this MinE-regulated dynamic assembly of MinD underlies MinD oscillation.
Figures
Figure 1
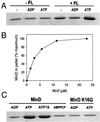
ATP regulates MinD binding to phospholipid vesicles. (A) MinD cosediments with phospholipid vesicles in the presence of ATP. Phospholipid vesicles (small unilamellar vesicles) were assembled from purified E. coli phospholipids by sonication. MinD (6 μM) was incubated with or without phospholipid vesicles (40 μg/ml) in the absence of nucleotide or with the addition of ADP or ATP. After 5 min incubation at 30°C the samples were centrifuged and the pellets were analyzed by SDS/PAGE on a 12.5% gel. (B) Saturable binding of MinD to phospholipid vesicles. MinD at various concentrations was incubated with phospholipid vesicles (40 μg/ml) and ATP and centrifuged, and the amount in the pellet was determined. The maximum amount of MinD in the pellet was set at 100%, which corresponded to 2.6 μM. (C) Analysis of MinD binding to membranes in the presence of the nonhydrolyzable analogues of ATP, ATPγS, and AMPCP. Also, the binding of MinD K16Q to phospholipid vesicles was determined. After incubation of MinD or MinD K16Q with phospholipid vesicles and nucleotide for 5 min at 30°C samples were centrifuged and the pellets were analyzed by SDS/PAGE.
Figure 2
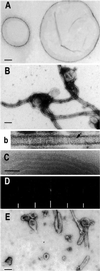
MinD tubulates vesicles in the presence of ATP. (A) Negative stain image of phospholipid vesicles. The diameter of the vesicles ranged from 0.1 to 2 μm. (B and b) Negative stain image of MinD tubes. A reaction mixture containing MinD (6 μM), phospholipid vesicles (40 μg/ml), and 1 mM ATP was incubated for 10 min at 30°C and samples were taken for EM and negative staining. The tube in b is at a higher magnification and striations are visible (see arrow). (C) Cryo-EM of MinD tubes. Tubes prepared as in B were visualized by cryo-EM. (D) The computed diffraction pattern of a tube visualized by cryo-EM indicates the tubes are well ordered and helical. The layer lines are indicated by hatch marks and are at 1/59 and 1/28.5 Å−1. Similar layer lines were obtained for tubes visualized by negative staining. (E) MinE causes disassembly of MinD tubes. MinE (6 μM) was added to MinD tubes prepared as in B and samples were taken 15 min later for EM. (Scale bars: 100 nm.)
Figure 3
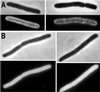
Localization of MinD K16Q in vivo. The localization of GFP-MinD was compared with GFP-MinD K16Q. (A) JS964 (Δ_min_) containing pZH106 (gfp_-minD) was induced for 2 h with 0.01% arabinose and photographed. Two cells are displayed. (B) JS964 (Δ_min) containing pZH106–16 (gfp-minD K16Q) was induced with 0.01% arabinose and examined by fluorescence microscopy 2 h later. Two cells are displayed. Immunoblot analysis demonstrated that the level of GFP-MinD and GFP-MinD K16Q were similar at 2 h after induction (data not shown).
Figure 4
MinE induces release of MinD from phospholipid vesicles. MinD (6 μM) was mixed with phospholipid (40 μg/ml) and 0.5 mM ATP to assemble tubes. MinE was added and incubated for 30 min. Samples were taken and the amount of MinD bound to membrane was determined by sedimentation and SDS/PAGE. In a second reaction ATPγS was used in place of ATP.
Figure 5
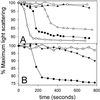
MinE induces tube disassembly as monitored by light scattering. Tubes were prepared as in Fig. 2_B_. A sample (300 μl) of the preassembled tubes was placed in a Hitachi fluorometer (model F-3010) and light scattering was monitored at 90°. (A) At 0 time MinE was added and the decrease in light scattering was measured. The MinE concentration was as follows: □, no MinE; ▴, 0.44 μM; ▵, 1.1 μM; ●, 2.2 μM; and ○, 4.4 μM. (B) □, no MinE; ♦, MinE (2.2 μM); ▾, MinE4 (2.2 μM) was added at 0 time to tubes assembled as in A and the change in light scattering was monitored.
Figure 6
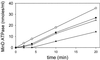
MinE stimulation of MinD ATPase activity displays a lag. MinD tubes were assembled as in Fig. 5_A_. At 0 time MinE was added and the ATPase activity was determined by measuring the release of 32Pi. The MinE concentrations are as follows: ▴, 0.44 μM; ▵, 1.1 μM; ●, 2.2 μM; and ○, 4.4 μM. The basal activity observed in the absence of MinE was subtracted from the values before plotting.
Figure 7
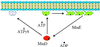
Model for dynamic association of MinD with the membrane. In this model MinD undergoes a conformational change upon binding ATP, causing it to associate with the membrane. Binding to the membrane causes an additional conformational change that favors the self-assembly of MinD into polymers. Interaction of MinD polymers with MinE leads to ATP hydrolysis and release of MinD. ATPγS is able to induce the first conformational change but MinD-ATPγS is unable to undergo the second change that would lead to assembly. MinD K16Q in the presence of ATP behaves similarly to MinD in the presence of ADP or AMPPCP (not diagrammed).
Similar articles
- ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro.
Lackner LL, Raskin DM, de Boer PA. Lackner LL, et al. J Bacteriol. 2003 Feb;185(3):735-49. doi: 10.1128/JB.185.3.735-749.2003. J Bacteriol. 2003. PMID: 12533449 Free PMC article. - Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE.
Suefuji K, Valluzzi R, RayChaudhuri D. Suefuji K, et al. Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):16776-81. doi: 10.1073/pnas.262671699. Epub 2002 Dec 13. Proc Natl Acad Sci U S A. 2002. PMID: 12482939 Free PMC article. - Bacterial cell division: a moving MinE sweeper boggles the MinD.
Margolin W. Margolin W. Curr Biol. 2001 May 15;11(10):R395-8. doi: 10.1016/s0960-9822(01)00217-2. Curr Biol. 2001. PMID: 11378404 Review. - MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation.
Lutkenhaus J, Sundaramoorthy M. Lutkenhaus J, et al. Mol Microbiol. 2003 Apr;48(2):295-303. doi: 10.1046/j.1365-2958.2003.03427.x. Mol Microbiol. 2003. PMID: 12675792 Review.
Cited by
- Mathematical model for positioning the FtsZ contractile ring in Escherichia coli.
Zhang Z, Morgan JJ, Lindahl PA. Zhang Z, et al. J Math Biol. 2014 Mar;68(4):911-30. doi: 10.1007/s00285-013-0652-z. Epub 2013 Feb 26. J Math Biol. 2014. PMID: 23440508 - Mechanism of the asymmetric activation of the MinD ATPase by MinE.
Park KT, Wu W, Lovell S, Lutkenhaus J. Park KT, et al. Mol Microbiol. 2012 Jul;85(2):271-81. doi: 10.1111/j.1365-2958.2012.08110.x. Epub 2012 Jun 7. Mol Microbiol. 2012. PMID: 22651575 Free PMC article. - Structures of the nucleoid occlusion protein SlmA bound to DNA and the C-terminal domain of the cytoskeletal protein FtsZ.
Schumacher MA, Zeng W. Schumacher MA, et al. Proc Natl Acad Sci U S A. 2016 May 3;113(18):4988-93. doi: 10.1073/pnas.1602327113. Epub 2016 Apr 18. Proc Natl Acad Sci U S A. 2016. PMID: 27091999 Free PMC article. - Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor.
Kang GB, Song HE, Kim MK, Youn HS, Lee JG, An JY, Chun JS, Jeon H, Eom SH. Kang GB, et al. Mol Microbiol. 2010 Jun 1;76(5):1222-31. doi: 10.1111/j.1365-2958.2010.07160.x. Epub 2010 Apr 14. Mol Microbiol. 2010. PMID: 20398219 Free PMC article. - Dynamic structures in Escherichia coli: spontaneous formation of MinE rings and MinD polar zones.
Huang KC, Meir Y, Wingreen NS. Huang KC, et al. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12724-8. doi: 10.1073/pnas.2135445100. Epub 2003 Oct 20. Proc Natl Acad Sci U S A. 2003. PMID: 14569005 Free PMC article.
References
- Yu X C, Margolin W. Mol Microbiol. 1999;32:315–326. - PubMed
- de Boer P A, Crossley R E, Rothfield L I. Cell. 1989;56:641–649. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases