Patterns of meiotic recombination in human fetal oocytes - PubMed (original) (raw)
. 2002 Jun;70(6):1469-79.
doi: 10.1086/340734. Epub 2002 May 1.
Affiliations
- PMID: 11992253
- PMCID: PMC379134
- DOI: 10.1086/340734
Patterns of meiotic recombination in human fetal oocytes
Charles Tease et al. Am J Hum Genet. 2002 Jun.
Abstract
Abnormal patterns of meiotic recombination (i.e., crossing-over) are believed to increase the risk of chromosome nondisjunction in human oocytes. To date, information on recombination has been obtained using indirect, genetic methods. Here we use an immunocytological approach, based on detection of foci of a DNA mismatch-repair protein, MLH1, on synaptonemal complexes at prophase I of meiosis, to provide the first direct estimate of the frequency of meiotic recombination in human oocytes. At pachytene, the stage of maximum homologous chromosome pairing, we found a mean of 70.3 foci (i.e., crossovers) per oocyte, with considerable intercell variability (range 48-102 foci). This mean equates to a genetic-map length of 3,515 cM. The numbers and positions of foci were determined for chromosomes 21, 18, 13, and X. These chromosomes yielded means of 1.23 foci (61.5 cM), 2.36 foci (118 cM), 2.5 foci (125 cM), and 3.22 foci (161 cM), respectively. The foci were almost invariably located interstitially and were only occasionally located close to chromosome ends. These data confirm the large difference, in recombination frequency, between human oocytes and spermatocytes and demonstrate a clear intersex variation in distribution of crossovers. In a few cells, chromosomes 21 and 18 did not have any foci (i.e., were presumptively noncrossover); however, configurations that lacked foci were not observed for chromosomes 13 and X. For the latter two chromosome pairs, the only instances of absence of foci were observed in abnormal cells that showed chromosome-pairing errors affecting these chromosomes. We speculate that these abnormal fetal oocytes may be the source of the nonrecombinant chromosomes 13 and X suggested, by genetic studies, to be associated with maternally derived chromosome nondisjunction.
Figures
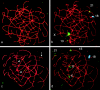
Figure 1
Paired images of two pachytene oocytes. SCs are shown in red, and MLH1 foci are shown in yellow. a and b, Oocyte with normal chromosome synapsis. The cell is shown after detection of MLH1 (a) and after FISH to identify selected chromosome pairs (b). Chromosome 21 has 1 focus, chromosome 18 has 3 foci, chromosome 13 is obscured by overlaps, and chromosome X has 5 foci. c and d, Abnormal oocyte showing synaptic errors. The cell is shown after MLH1 detection (c) and subsequent FISH (d). Chromosome X displays synaptic failure and, as a result, is present as two separate AEs (leftward-pointing arrowheads); chromosome 18 shows partial synapsis (downward-pointing arrow). MLH1 foci are present along fully synapsed bivalents and on the synapsed segment of chromosome 18 (c); there are no foci on the asynapsed axes of chromosomes X.
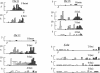
Figure 2
Histograms showing the distributions of foci. Each chromosome is divided into 5% intervals (see “Material and Methods”), and the position of the centromere is indicated by a blackened oval on the _X-_axis. The positions of foci are shown separately (for bivalents with 1, 2, 3, etc., foci). For ease of presentation, the chromosome pairs are not shown to scale.
Similar articles
- Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells.
Tease C, Hultén MA. Tease C, et al. Cytogenet Genome Res. 2004;107(3-4):208-15. doi: 10.1159/000080599. Cytogenet Genome Res. 2004. PMID: 15467366 - Altered patterns of meiotic recombination in human fetal oocytes with asynapsis and/or synaptonemal complex fragmentation at pachytene.
Tease C, Hartshorne G, Hultén M. Tease C, et al. Reprod Biomed Online. 2006 Jul;13(1):88-95. doi: 10.1016/s1472-6483(10)62020-2. Reprod Biomed Online. 2006. PMID: 16820117 - Discontinuities and unsynapsed regions in meiotic chromosomes have a cis effect on meiotic recombination patterns in normal human males.
Sun F, Oliver-Bonet M, Liehr T, Starke H, Trpkov K, Ko E, Rademaker A, Martin RH. Sun F, et al. Hum Mol Genet. 2005 Oct 15;14(20):3013-8. doi: 10.1093/hmg/ddi332. Epub 2005 Sep 9. Hum Mol Genet. 2005. PMID: 16155114 - Female Meiosis: Synapsis, Recombination, and Segregation in Drosophila melanogaster.
Hughes SE, Miller DE, Miller AL, Hawley RS. Hughes SE, et al. Genetics. 2018 Mar;208(3):875-908. doi: 10.1534/genetics.117.300081. Genetics. 2018. PMID: 29487146 Free PMC article. Review. - [Homologs of MutS and MutL during mammalian meiosis].
Santucci-Darmanin S, Paquis-Flucklinger V. Santucci-Darmanin S, et al. Med Sci (Paris). 2003 Jan;19(1):85-91. doi: 10.1051/medsci/200319185. Med Sci (Paris). 2003. PMID: 12836196 Review. French.
Cited by
- High-resolution crossover maps for each bivalent of Zea mays using recombination nodules.
Anderson LK, Doyle GG, Brigham B, Carter J, Hooker KD, Lai A, Rice M, Stack SM. Anderson LK, et al. Genetics. 2003 Oct;165(2):849-65. doi: 10.1093/genetics/165.2.849. Genetics. 2003. PMID: 14573493 Free PMC article. - Association of ovarian stimulation and embryonic aneuploidy in in vitro fertilization cycles with preimplantation genetic testing: A narrative systematic review.
Rodriguez-Purata J, Gomez-Cuesta MJ, Cervantes-Bravo E. Rodriguez-Purata J, et al. JBRA Assist Reprod. 2022 Apr 17;26(2):348-361. doi: 10.5935/1518-0557.20210069. JBRA Assist Reprod. 2022. PMID: 34751016 Free PMC article. - Shugoshin protects centromere pairing and promotes segregation of nonexchange partner chromosomes in meiosis.
Previato de Almeida L, Evatt JM, Chuong HH, Kurdzo EL, Eyster CA, Gladstone MN, Gómez-H L, Llano E, Meyer R, Pendas AM, Pezza RJ, Dawson DS. Previato de Almeida L, et al. Proc Natl Acad Sci U S A. 2019 May 7;116(19):9417-9422. doi: 10.1073/pnas.1902526116. Epub 2019 Apr 24. Proc Natl Acad Sci U S A. 2019. PMID: 31019073 Free PMC article. - Reduced meiotic recombination on the XY bivalent is correlated with an increased incidence of sex chromosome aneuploidy in men with non-obstructive azoospermia.
Sun F, Mikhaail-Philips M, Oliver-Bonet M, Ko E, Rademaker A, Turek P, Martin RH. Sun F, et al. Mol Hum Reprod. 2008 Jul;14(7):399-404. doi: 10.1093/molehr/gan030. Epub 2008 Jun 26. Mol Hum Reprod. 2008. PMID: 18583429 Free PMC article. - Analysis of clinical outcomes and meiotic segregation modes following preimplantation genetic testing for structural rearrangements using aCGH/NGS in couples with balanced chromosome rearrangement.
Nakano T, Ammae M, Satoh M, Mizuno S, Nakaoka Y, Morimoto Y. Nakano T, et al. Reprod Med Biol. 2022 Jun 29;21(1):e12476. doi: 10.1002/rmb2.12476. eCollection 2022 Jan-Dec. Reprod Med Biol. 2022. PMID: 35781920 Free PMC article.
References
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 13:336–342 - PubMed
- Barlow AL, Hultén MA (1998) Crossing over analysis at pachytene in man. Eur J Hum Genet 6:350–358 - PubMed
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6:989–998 - PubMed
- Bojko M (1983) Human meiosis. VIII. Chromosome pairing and formation of the synaptonemal complex in oocytes. Carlsberg Res Commun 48:457–483
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources