Genomewide comparison of DNA sequences between humans and chimpanzees - PubMed (original) (raw)
Comparative Study
doi: 10.1086/340787. Epub 2002 Apr 30.
Affiliations
- PMID: 11992255
- PMCID: PMC379137
- DOI: 10.1086/340787
Comparative Study
Genomewide comparison of DNA sequences between humans and chimpanzees
Ingo Ebersberger et al. Am J Hum Genet. 2002 Jun.
Abstract
A total of 8,859 DNA sequences encompassing approximately 1.9 million base pairs of the chimpanzee genome were sequenced and compared to corresponding human DNA sequences. Although the average sequence difference is low (1.24%), the extent of changes is markedly different among sites and types of substitutions. Whereas approximately 15% of all CpG sites have experienced changes between humans and chimpanzees, owing to a 23-fold excess of transitions and a 7-fold excess of transversions, substitutions at other sites vary in frequency, between 0.1% and 0.5%. If the nucleotide diversity in the common ancestral species of humans and chimpanzees is assumed to have been about fourfold higher than in contemporary humans, all possible comparisons between autosomes and X and Y chromosomes result in estimates of the ratio between male and female mutation rates of approximately 3. Thus, the relative time spent in the male and female germlines may be a major determinant of the overall accumulation of nucleotide substitutions. However, since the extent of divergence differs significantly among autosomes, additional unknown factors must also influence the accumulation of substitutions in the human genome.
Figures
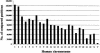
Figure 1
Histogram showing the number of DNA sequence positions compared per human chromosome
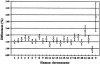
Figure 2
Mean DNA sequence differences between humans and chimpanzees by human chromosome. Bars indicate 95% CIs.
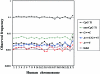
Figure 3
Frequency distribution of DNA sequence differences between chimpanzees and humans, for each human chromosome. Ti = transitions.
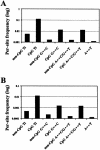
Figure 4
Genomewide average frequencies for various nucleotide differences between chimpanzees and humans (A) and among humans (B). Ti = transitions.
Similar articles
- Mutation rates in humans. II. Sporadic mutation-specific rates and rate of detrimental human mutations inferred from hemophilia B.
Giannelli F, Anagnostopoulos T, Green PM. Giannelli F, et al. Am J Hum Genet. 1999 Dec;65(6):1580-7. doi: 10.1086/302652. Am J Hum Genet. 1999. PMID: 10577911 Free PMC article. - Strong and weak male mutation bias at different sites in the primate genomes: insights from the human-chimpanzee comparison.
Taylor J, Tyekucheva S, Zody M, Chiaromonte F, Makova KD. Taylor J, et al. Mol Biol Evol. 2006 Mar;23(3):565-73. doi: 10.1093/molbev/msj060. Epub 2005 Nov 9. Mol Biol Evol. 2006. PMID: 16280537 - Evidence for a complex demographic history of chimpanzees.
Fischer A, Wiebe V, Pääbo S, Przeworski M. Fischer A, et al. Mol Biol Evol. 2004 May;21(5):799-808. doi: 10.1093/molbev/msh083. Epub 2004 Feb 12. Mol Biol Evol. 2004. PMID: 14963091 - Understanding the recent evolution of the human genome: insights from human-chimpanzee genome comparisons.
Kehrer-Sawatzki H, Cooper DN. Kehrer-Sawatzki H, et al. Hum Mutat. 2007 Feb;28(2):99-130. doi: 10.1002/humu.20420. Hum Mutat. 2007. PMID: 17024666 Review. - A map of the common chimpanzee genome.
Wildman DE. Wildman DE. Bioessays. 2002 Jun;24(6):490-3. doi: 10.1002/bies.10103. Bioessays. 2002. PMID: 12111730 Review.
Cited by
- Combining DNA and protein alignments to improve genome annotation with LiftOn.
Chao KH, Heinz JM, Hoh C, Mao A, Shumate A, Pertea M, Salzberg SL. Chao KH, et al. bioRxiv [Preprint]. 2024 May 17:2024.05.16.593026. doi: 10.1101/2024.05.16.593026. bioRxiv. 2024. PMID: 38798552 Free PMC article. Preprint. - Distinct evolutionary lineages of Schistocephalus parasites infecting co-occurring sculpin and stickleback fishes in Alaska.
Heins DC, Moody KN, Arostegui MC, Harmon BS, Blum MJ, Quinn TP. Heins DC, et al. Parasitology. 2024 May;151(6):626-633. doi: 10.1017/S0031182024000593. Epub 2024 May 9. Parasitology. 2024. PMID: 38719483 Free PMC article. - Whole-genome resource sequences of 57 indigenous Ethiopian goats.
Belay S, Belay G, Nigussie H, Jian-Lin H, Tijjani A, Ahbara AM, Tarekegn GM, Woldekiros HS, Mor S, Dobney K, Lebrasseur O, Hanotte O, Mwacharo JM. Belay S, et al. Sci Data. 2024 Jan 29;11(1):139. doi: 10.1038/s41597-024-02973-2. Sci Data. 2024. PMID: 38287052 Free PMC article. - Evolutionary dynamics of pseudoautosomal region 1 in humans and great apes.
Bergman J, Schierup MH. Bergman J, et al. Genome Biol. 2022 Oct 17;23(1):215. doi: 10.1186/s13059-022-02784-x. Genome Biol. 2022. PMID: 36253794 Free PMC article.
References
Electronic-Database Information
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- Genome Software Development Page, http://www.phrap.org/
- Human Genome Working Draft, http://genome.ucsc.edu/
- Repeat Masker Server Home Page, http://repeatmasker.genome.washington.edu/
- UCSC Genome Browser Gateway, http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg10
References
- Bailey WJ, Fitch DH, Tagle DA, Czelusniak J, Slightom JL, Goodman M (1991) Molecular evolution of the psi eta-globin gene locus: gibbon phylogeny and the hominoid slowdown. Mol Biol Evol 8:155–184 - PubMed
- Bernardi G (1995) The human genome: organization and evolutionary history. Annu Rev Genet 29:445–476 - PubMed
- Bohossian HB, Skaletsky H, Page DC (2000) Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature 406:622–625 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources