Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus - PubMed (original) (raw)
. 2002 May 14;99(10):6573-8.
doi: 10.1073/pnas.092569199. Epub 2002 May 7.
Duc Bui Minh, Tamás Henics, Agnieszka Dryla, Birgit Winkler, Christine Triska, Aoife P Boyd, Johannes Söllner, Walter Schmidt, Uwe von Ahsen, Michael Buschle, Steven R Gill, James Kolonay, Hanif Khalak, Claire M Fraser, Alexander von Gabain, Eszter Nagy, Andreas Meinke
Affiliations
- PMID: 11997460
- PMCID: PMC124444
- DOI: 10.1073/pnas.092569199
Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus
Hildegard Etz et al. Proc Natl Acad Sci U S A. 2002.
Abstract
For the design of potent subunit vaccines, it is of paramount importance to identify all antigens immunologically recognized by a patient population infected with a pathogen. We have developed a rapid and efficient procedure to identify such commonly recognized antigens, and here we provide a comprehensive in vivo antigenic profile of Staphylococcus aureus, an important human pathogen. S. aureus peptides were displayed on the surface of Escherichia coli via fusion to one of two outer membrane proteins (LamB and FhuA) and probed with sera selected for high Ab titer and opsonic activity. A total of 60 antigenic proteins were identified, most of which are located or predicted to be located on the surface of the bacterium or secreted. The identification of these antigens and their reactivity with individual sera from patients and healthy individuals greatly facilitate the selection of promising vaccine candidates for further evaluation. This approach, which makes use of whole genome sequence information, has the potential to greatly accelerate and facilitate the formulation of novel vaccines and is applicable to any pathogen that induces Abs in humans and/or experimental animals.
Figures
Figure 1
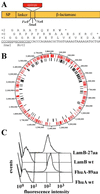
Surface display of peptides derived from random genomic fragments by E. coli. (A) Genomic fragments were frame-selected by using the β-lactamase selection vector pMAL4.1. Insertion of fragments into the Sma_I site fulfilling the 3_n + 1 length requirement restore the reading frame of β-lactamase, whereas the other two reading frames (3_n_ + 0, 3_n_ + 2) result in premature termination. SP, signal peptide. (B) The sequence of 600 randomly picked clones from the LSA50 library was determined and aligned with the chromosome of S. aureus. Red and black bars correspond to matches to ORFs and to noncoding sequences, respectively. (C) Efficient surface display of the T7⋅tag in loop 5 of LamB (27 aa) and of six myc epitopes in loop 5 of FhuA (89 aa) is shown by FACS analysis. Cells were stained with mAb T7⋅Tag (Novagen) and mAb α-myc 9E10, respectively.
Figure 2
Characterization of human serum collections. (A) Cumulative Ab levels were determined by ELISA by using multiple S. aureus antigens. Antistaphylococcal Ab levels were measured individually with total S. aureus lysate, lipoteichoic acid, peptidoglycan, and 13 recombinant proteins: clumping factor A and B (ClfA, ClfB), Fibronectin-binding protein (FnBPA), SD-repeat proteins (SdrC, SdrE), MHC class II analogous protein (Map-w), elastin-binding protein, enolase, iron transport lipoproteins (LP309, LP342), Sortase, Coagulase, and extracellular fibrinogen-binding protein. Individual sera from healthy individuals were ranked on the basis of their IgG titer against each antigen. The total score was calculated as a sum of the ranking scores against individual antigens. The selection of patient and normal sera was based on the same criteria. (B) IgGs from five pooled high-titer healthy normal (N1) or patient (P1) sera were tested for opsonic activity in an in vitro opsonophagocytosis assay. FITC-labeled 8325–4 protein A deficient S. aureus cells (107) were preopsonized with 100 μg of human IgGs in the absence or presence of complement (C′). Bacteria were incubated with 106 freshly isolated human polymorphonuclear cells (PMNs), and opsonophagocytosis was measured by FACS analysis. (C) Control: PMNs incubated with labeled bacteria in the absence of IgG or C′.
Figure 3
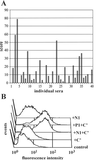
Identification of antigenic proteins from S. aureus by MACS. (A) The LSA50-LamB library was screened with an IgG preparation from patients (P1) by MACS selection. Forty-eight randomly picked clones were tested with patient serum P1 (1 μg/ml IgG) and anti-LamB mAbs at a dilution of 1:2,000. (B) Epitopes recovered from the LSA50-LamB screen (black bars) and the LSA250-FhuA screen (gray bars) with P1 serum and aligned with the sequence of the gene encoding Empbp are localized mainly in two regions. Numbers next to bars represent the frequency of recovery of these epitopes. (C) Comparison of the number of proteins identified in screens with human patient serum in the two platforms LamB and FhuA and of those from screens performed with patient sera and high-titer sera from healthy individuals.
Figure 4
Evaluation of antigenic epitopes. (A) Reactivities of a selection of 10 individual patient sera and patient pool P1 with 9 identified epitopes. The location of the epitope within the protein is shown schematically. (B) Approximately 109 lysozyme-treated and sonicated bacteria expressing FhuA or a fusion of the antigenic epitope and FhuA (e.g., FhuA-209) were injected intravenously into BALB/c mice. The four epitopes embedded in FhuA were expressed at comparable levels in E. coli on the basis of Western blot analysis with polyclonal anti-FhuA Abs. Serum was taken 2 wk after the first and second injections and depleted for Abs directed against E. coli or FhuA protein with a cellular lysate from E. coli expressing wild-type FhuA. FhuA fusion proteins were detected by Western blot analysis in lysates from an equivalent of 5 × 106 bacteria. Sera from mice injected with FhuA-858, 209, and 507 were diluted 1:5,000, sera from animals injected with FhuA-723 were diluted 1:50,000, and preimmune serum 1:1,000. Horseradish peroxidase (HRP)-coupled anti-mouse Igs were used at a dilution of 1:5,000. (C) The same lysate as above was assessed for reactivity with IgG type-specific Abs by using biotinylated anti-mouse IgG1 and IgG2a as secondary Abs at a concentration of 0.2 and 0.05 μg/ml, respectively. HRP-coupled streptavidin was diluted 1:5,000.
Figure 5
Opsonization of S. aureus with B cell epitope-specific human Abs. IgGs were purified from human sera by affinity chromatography by using a 15-aa-long synthetic peptide (PPKDTNQTQPATQPA) representing one LPXTGp5 epitope. Alexa-Fluor-labeled S. aureus Wood 46 (STAW) cells (107) were preopsonized with 0.5 μg of epitope-specific anti-LPXTGp5 Ab in the presence of 5 μg of LPXTGp5-derived specific (epitope) or nonspecific (nonepitope) peptides. Uptake was registered as a percentage of phagocytic P388 mouse monocytic cells (106) with increased fluorescence after 15-min incubation with labeled STAW measured by FACS. The control sample represents P388 cells incubated with labeled bacteria in the absence of complement (C′).
Similar articles
- DNA-protein immunization against the GapB and GapC proteins of a mastitis isolate of Staphylococcus aureus.
Kerro-Dego O, Prysliak T, Potter AA, Perez-Casal J. Kerro-Dego O, et al. Vet Immunol Immunopathol. 2006 Sep 15;113(1-2):125-38. doi: 10.1016/j.vetimm.2006.04.004. Epub 2006 Jun 13. Vet Immunol Immunopathol. 2006. PMID: 16777237 - Immune responses to a Staphylococcus aureus GapC/B chimera and its potential use as a component of a vaccine for S. aureus mastitis.
Perez-Casal J, Prysliak T, Kerro-Dego O, Potter AA. Perez-Casal J, et al. Vet Immunol Immunopathol. 2006 Jan 15;109(1-2):85-97. doi: 10.1016/j.vetimm.2005.07.024. Epub 2005 Sep 13. Vet Immunol Immunopathol. 2006. PMID: 16165220 - An immunogenicity study of a newly fusion protein Cna-FnBP vaccinated against Staphylococcus aureus infections in a mice model.
Zhou H, Xiong ZY, Li HP, Zheng YL, Jiang YQ. Zhou H, et al. Vaccine. 2006 May 29;24(22):4830-7. doi: 10.1016/j.vaccine.2006.03.020. Epub 2006 Mar 24. Vaccine. 2006. PMID: 16616974 - Staphylococcus aureus antigens and challenges in vaccine development.
Middleton JR. Middleton JR. Expert Rev Vaccines. 2008 Aug;7(6):805-15. doi: 10.1586/14760584.7.6.805. Expert Rev Vaccines. 2008. PMID: 18665778 Review. - Towards the immune proteome of Staphylococcus aureus - The anti-S. aureus antibody response.
Holtfreter S, Kolata J, Bröker BM. Holtfreter S, et al. Int J Med Microbiol. 2010 Feb;300(2-3):176-92. doi: 10.1016/j.ijmm.2009.10.002. Epub 2009 Nov 3. Int J Med Microbiol. 2010. PMID: 19889576 Review.
Cited by
- A comprehensive comparison of DNA and RNA vaccines.
Wang C, Yuan F. Wang C, et al. Adv Drug Deliv Rev. 2024 Jul;210:115340. doi: 10.1016/j.addr.2024.115340. Epub 2024 May 27. Adv Drug Deliv Rev. 2024. PMID: 38810703 Review. - Vaccines and Immunoinformatics for Vaccine Design.
Joon S, Singla RK, Shen B. Joon S, et al. Adv Exp Med Biol. 2022;1368:95-110. doi: 10.1007/978-981-16-8969-7_5. Adv Exp Med Biol. 2022. PMID: 35594022 - Natural Human Immunity Against Staphylococcal Protein A Relies on Effector Functions Triggered by IgG3.
Boero E, Cruz AR, Pansegrau W, Giovani C, Rooijakkers SHM, van Kessel KPM, van Strijp JAG, Bagnoli F, Manetti AGO. Boero E, et al. Front Immunol. 2022 Mar 11;13:834711. doi: 10.3389/fimmu.2022.834711. eCollection 2022. Front Immunol. 2022. PMID: 35359919 Free PMC article. - Advanced strategies for development of vaccines against human bacterial pathogens.
Sharma A, Sanduja P, Anand A, Mahajan P, Guzman CA, Yadav P, Awasthi A, Hanski E, Dua M, Johri AK. Sharma A, et al. World J Microbiol Biotechnol. 2021 Mar 22;37(4):67. doi: 10.1007/s11274-021-03021-6. World J Microbiol Biotechnol. 2021. PMID: 33748926 Free PMC article. Review. - Subdominance in Antibody Responses: Implications for Vaccine Development.
Lindahl G. Lindahl G. Microbiol Mol Biol Rev. 2020 Nov 25;85(1):e00078-20. doi: 10.1128/MMBR.00078-20. Print 2020 Nov 25. Microbiol Mol Biol Rev. 2020. PMID: 33239435 Free PMC article. Review.
References
- Pelton S I. Vaccine. 2000;19:S96–S99. - PubMed
- Pizza M, Scarlato V, Masignani V, Giuliani M M, Arico B, Comanducci M, Jennings G T, Baldi L, Bartolini E, Capecchi B, et al. Science. 2000;287:1816–1820. - PubMed
- Chakravarti D N, Fiske M J, Fletcher L D, Zagursky R J. Vaccine. 2000;19:601–612. - PubMed
- Jungblut P R, Schaible U E, Mollenkopf H J, Zimny-Arndt U, Raupach B, Mattow J, Halada P, Lamer S, Hagens K, Kaufmann S H, et al. Mol Microbiol. 1999;33:1103–1117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical