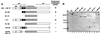The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization - PubMed (original) (raw)
The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization
Karim Bouazoune et al. EMBO J. 2002.
Abstract
Drosophila Mi-2 (dMi-2) is the ATPase subunit of a complex combining ATP-dependent nucleosome remodelling and histone deacetylase activities. dMi-2 contains an HMG box-like region, two PHD fingers, two chromodomains and a SNF2-type ATPase domain. It is not known which of these domains contribute to nucleosome remodelling. We have tested a panel of dMi-2 deletion mutants in ATPase, nucleosome mobilization and nucleosome binding assays. Deletion of the chromodomains impairs all three activities. A dMi-2 mutant lacking the chromodomains is incorporated into a functional histone deacetylase complex in vivo but has lost nucleosome-stimulated ATPase activity. In contrast to dHP1, dMi-2 does not bind methylated histone H3 tails and does not require histone tails for nucleosome binding. Instead, the dMi-2 chromodomains display DNA binding activity that is not shared by other chromodomains. Our results suggest that the chromodomains act at an early step of the remodelling process to bind the nucleosome substrate predominantly via protein-DNA interactions. Furthermore, we identify DNA binding as a novel chromodomain-associated activity.
Figures
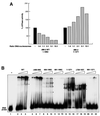
Fig. 1. The dMi-2 chromodomain region is important for nucleosome mobilization. (A) Schematic representation of dMi-2 mutants used. Domains are indicated on top. HMG, HMG box-like region; PHD, PHD fingers; chromo, chromodomains; NTR, N-terminal region; CTR, C-terminal region; WT, wild type. (B) Coomassie Blue-stained SDS polyacrylamide gel of recombinant dMi-2 mutants. Three hundred (odd-numbered lanes) and 600 ng (even-numbered lanes) of each protein were loaded as indicated. MW, molecular weight marker. (C) Nucleosome mobilization assay. Increasing amounts of recombinant dMi-2 proteins were incubated with a radioactively labelled end-positioned 248 bp nucleosome in the absence (–) or presence (+) of ATP as indicated. Nucleosomes were resolved by native PAGE. Positions of endpositioned (open circle) and centrally positioned (filled circle) nucleosomes are indicated.
Fig. 1. The dMi-2 chromodomain region is important for nucleosome mobilization. (A) Schematic representation of dMi-2 mutants used. Domains are indicated on top. HMG, HMG box-like region; PHD, PHD fingers; chromo, chromodomains; NTR, N-terminal region; CTR, C-terminal region; WT, wild type. (B) Coomassie Blue-stained SDS polyacrylamide gel of recombinant dMi-2 mutants. Three hundred (odd-numbered lanes) and 600 ng (even-numbered lanes) of each protein were loaded as indicated. MW, molecular weight marker. (C) Nucleosome mobilization assay. Increasing amounts of recombinant dMi-2 proteins were incubated with a radioactively labelled end-positioned 248 bp nucleosome in the absence (–) or presence (+) of ATP as indicated. Nucleosomes were resolved by native PAGE. Positions of endpositioned (open circle) and centrally positioned (filled circle) nucleosomes are indicated.
Fig. 2. ATPase activity and nucleosome binding of recombinant dMi-2 proteins. (A) Recombinant dMi-2 (90 fmol) was incubated with γ-32P-labelled ATP in the presence of 100 ng DNA or 100 ng nucleosomes as indicated. ATP hydrolysis was detected by thin-layer chromatography and quantified by phosphoimager analysis. Activity is plotted relative to the nucleosome-stimulated ATPase activity of wild-type dMi-2, which is set to 100. The data presented are derived from three independent experiments. (B) Recombinant dMi-2 proteins (150, 300, 450 fmol) were incubated with a radioactively labelled 146 bp mononucleosome as indicated. Complexes were resolved by native PAGE. Empty circles denote positions of dMi-2–nucleosome complexes.
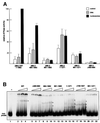
Fig. 3. Chromodomains are required for nucleosome-stimulated ATPase activity of the dMi-2 complex. (A) Limited proteolysis. One microgram of recombinant wild-type dMi-2 (lanes 1–3) or dMi-2 ΔC (lanes 4–6) were digested with increasing amounts of trypsin as indicated. Tryptic fragments were visualized by SDS–PAGE and western blotting using αdMi-2-specific antiserum (αdMi2-C). As a control, recombinant dMi-2 691–1271 (ATPase domain) was loaded in lane 7. Molecular weights are shown on the left. (B) GST pull-down. GST (lanes 2, 5 and 8) or a GST–dRPD3 fusion (lanes 3, 6 and 9) immobilized on beads were incubated with 35S-labelled in vitro translated (IVT) luciferase, dMi-2 WT and dMi-2 ΔC as indicated on top of the figure. Beads were washed extensively and bound material was visualized by SDS–PAGE and autoradiography. In vitro translated proteins were loaded as an input control (10%; lanes 1, 4 and 7). Positions of dMi-2 WT, dMi-2 ΔC and luciferase are indicated by arrows. (C) Co-infection/co-immunoprecipitation. Sf9 cells were co-infected with different combinations of recombinant baculoviruses as indicated on the top of the panels. Extracts from infected cells were immunoprecipitated with immobilized α-flag antibody, beads were extensively washed, subjected to SDS–PAGE and western analysis using specific antibodies as indicated on the right of the panels (lanes IP). To control for expression from the recombinant baculoviruses, an aliquot of cell extracts was loaded for comparison (lanes IN). (D) The dMi-2 chromodomain region is not required for formation of dMi-2 histone deacetylase complexes in vivo. Whole-cell extracts from SL2 cell lines stably expressing flag-tagged dMi-2 (SL2-WT), flag-tagged dMi-2 Δ484–691 (SL2-Δchromo) and from a control SL2 line (SL2-0) were fractionated by cation exchange chromatography. The dMi-2 containing fraction (500 mM KCl eluate) was subjected to affinity purification with immobilized α-flag antibody. Flag-tagged complexes were eluted with an excess of flag peptide. Eluted complexes were subjected to western analysis using antisera directed against dMi-2 (upper panel), dRPD3 (middle panel) and p55 (lower panel). (E) Eluted complexes were assayed for histone deacetylase activity. One and 2 µl of each complex preparation were used, as indicated. (F) Eluted complexes were subjected to ATPase assays in the presence or absence of 100 ng DNA or nucleosomes, as indicated. ATP hydrolysis was detected and quantified as above.
Fig. 3. Chromodomains are required for nucleosome-stimulated ATPase activity of the dMi-2 complex. (A) Limited proteolysis. One microgram of recombinant wild-type dMi-2 (lanes 1–3) or dMi-2 ΔC (lanes 4–6) were digested with increasing amounts of trypsin as indicated. Tryptic fragments were visualized by SDS–PAGE and western blotting using αdMi-2-specific antiserum (αdMi2-C). As a control, recombinant dMi-2 691–1271 (ATPase domain) was loaded in lane 7. Molecular weights are shown on the left. (B) GST pull-down. GST (lanes 2, 5 and 8) or a GST–dRPD3 fusion (lanes 3, 6 and 9) immobilized on beads were incubated with 35S-labelled in vitro translated (IVT) luciferase, dMi-2 WT and dMi-2 ΔC as indicated on top of the figure. Beads were washed extensively and bound material was visualized by SDS–PAGE and autoradiography. In vitro translated proteins were loaded as an input control (10%; lanes 1, 4 and 7). Positions of dMi-2 WT, dMi-2 ΔC and luciferase are indicated by arrows. (C) Co-infection/co-immunoprecipitation. Sf9 cells were co-infected with different combinations of recombinant baculoviruses as indicated on the top of the panels. Extracts from infected cells were immunoprecipitated with immobilized α-flag antibody, beads were extensively washed, subjected to SDS–PAGE and western analysis using specific antibodies as indicated on the right of the panels (lanes IP). To control for expression from the recombinant baculoviruses, an aliquot of cell extracts was loaded for comparison (lanes IN). (D) The dMi-2 chromodomain region is not required for formation of dMi-2 histone deacetylase complexes in vivo. Whole-cell extracts from SL2 cell lines stably expressing flag-tagged dMi-2 (SL2-WT), flag-tagged dMi-2 Δ484–691 (SL2-Δchromo) and from a control SL2 line (SL2-0) were fractionated by cation exchange chromatography. The dMi-2 containing fraction (500 mM KCl eluate) was subjected to affinity purification with immobilized α-flag antibody. Flag-tagged complexes were eluted with an excess of flag peptide. Eluted complexes were subjected to western analysis using antisera directed against dMi-2 (upper panel), dRPD3 (middle panel) and p55 (lower panel). (E) Eluted complexes were assayed for histone deacetylase activity. One and 2 µl of each complex preparation were used, as indicated. (F) Eluted complexes were subjected to ATPase assays in the presence or absence of 100 ng DNA or nucleosomes, as indicated. ATP hydrolysis was detected and quantified as above.
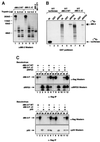
Fig. 4. dMi-2 does not require histone tails for nucleosome binding. (A) Drosophila embryo nuclear extract was incubated with immobilized methylated and unmethylated histone H3 peptides as indicated. Peptide beads were successively washed with buffer containing increasing concentration of NaCl and eluted with glycine buffer pH 2.5, as indicated on top of the panels. Proteins in unbound, wash and eluate fractions were detected by western analysis using antisera directed against dHP1 (upper panel) and dMi-2 (lower panel). IN, input; control, unmodified peptide; K9me, K9-methylated peptide; K4me, K4-methylated peptide. (B) dMi-2 nucleosome bandshift assay using a 146 bp nucleosome probe reconstituted from intact recombinant histones (lanes 1–6) or reconstituted from tailless recombinant histones (lanes 7–12). Increasing amounts of recombinant dMi-2 WT (lanes 2 and 8, 50 fmol; lanes 3 and 9, 100 fmol; lanes 4 and 10, 200 fmol; lanes 5 and 11, 300 fmol; lanes 6 and 12, 400 fmol) were incubated with radioactively labelled nucleosome probe. Complexes were resolved by native PAGE. The positions of free nucleosome probe, dMi-2–nucleosome complexes and nucleosome probe precipitated in the loading well are indicated.
Fig. 5. The chromodomain region of dMi-2 is required for DNA binding. (A) dMi-2 was pre-incubated with DNA prior to addition of 100 ng nucleosomes and ATP at different DNA:nucleosome ratios, as indicated. ATPase activity was determined as above and expressed as percentage nucleosome-stimulated ATPase activity. (B) dMi-2 proteins (40, 80 and 160 fmol) were incubated with a radioactively labelled 146 bp DNA fragment, as indicated. Complexes were resolved by native PAGE. Empty circles denote the position of complexes. (C) dMi-2 proteins (40, 80 and 160 fmol) were subjected to the bandshift assay using a 146 bp DNA fragment (upper panel) or a 146 bp nucleosome (lower panel) as a probe as indicated. Complexes are denoted by arrowheads.
Fig. 5. The chromodomain region of dMi-2 is required for DNA binding. (A) dMi-2 was pre-incubated with DNA prior to addition of 100 ng nucleosomes and ATP at different DNA:nucleosome ratios, as indicated. ATPase activity was determined as above and expressed as percentage nucleosome-stimulated ATPase activity. (B) dMi-2 proteins (40, 80 and 160 fmol) were incubated with a radioactively labelled 146 bp DNA fragment, as indicated. Complexes were resolved by native PAGE. Empty circles denote the position of complexes. (C) dMi-2 proteins (40, 80 and 160 fmol) were subjected to the bandshift assay using a 146 bp DNA fragment (upper panel) or a 146 bp nucleosome (lower panel) as a probe as indicated. Complexes are denoted by arrowheads.
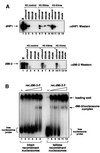
Fig. 6. Chromodomains and DNA binding. (A) DNA and nucleosome bandshift assay using bacterially expressed dMi-2 chromodomain polypeptides. As a control, material obtained from a mock purification from E.coli transformed with empty expression vector was used (‘control’, lanes 3–8). c1+2, dMi-2 488–673; c1, dMi-2 488–566; c2, dMi-2 608–673. The amounts of chromodomain peptides used were as follows: lanes 9, 14, 15, 20, 21 and 26: 6 pmol; lanes 10, 13, 16, 19, 22 and 25: 12 pmol; lanes 11, 12, 17, 18, 23 and 24: 25 pmol. Complexes are denoted with open (chromodomain–DNA complexes) and closed (chromodomain–nucleosome complexes) arrowheads. (B) DNA bandshift assay using bacterially expressed chromodomain polypeptides. Position of chromodomain–DNA complexes are shown by open arrowheads. The amounts of chromodomain peptides used were as follows: lanes 2, 5, 8, 12 and 16: 6 pmol; lanes 3, 6, 9, 13 and 17: 12 pmol; lanes 4, 7, 10, 14 and 18: 25 pmol; lanes 11, 15 and 19: 50 pmol. Chromodomain–DNA complexes are denoted with open arrowheads.
Fig. 7. Alignment of chromodomains. The chromodomain sequences of Drosophila HP1 (Dm_HP1; aa residues 23–74), Drosophila Polycomb (Dm_Pc) and the two dMi-2 chromodomains (Dm_dMi-2 c1 and Dm_dMi-2 c2) were aligned. Conserved residues are shaded. Positions of α-helix, β-sheets and loops are indicated below the alignment according to the HP1 structure as determined by Ball et al. (1997). HP1 residues forming the hydrophobic core of the structure are denoted with open circles above of the alignment. HP1 residues involved in methylated histone tail recognition are indictated by filled circles (Jacobs et al., 2001).
Similar articles
- dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties.
Brehm A, Längst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Müller J, Becker PB. Brehm A, et al. EMBO J. 2000 Aug 15;19(16):4332-41. doi: 10.1093/emboj/19.16.4332. EMBO J. 2000. PMID: 10944116 Free PMC article. - Double chromodomains cooperate to recognize the methylated histone H3 tail.
Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Flanagan JF, et al. Nature. 2005 Dec 22;438(7071):1181-5. doi: 10.1038/nature04290. Nature. 2005. PMID: 16372014 - Mi-2 chromatin remodeling factor functions in sensory organ development through proneural gene repression in Drosophila.
Yamasaki Y, Nishida Y. Yamasaki Y, et al. Dev Growth Differ. 2006 Sep;48(7):411-8. doi: 10.1111/j.1440-169X.2006.00880.x. Dev Growth Differ. 2006. PMID: 16961588 - Chromatin remodeling by ATP-dependent molecular machines.
Lusser A, Kadonaga JT. Lusser A, et al. Bioessays. 2003 Dec;25(12):1192-200. doi: 10.1002/bies.10359. Bioessays. 2003. PMID: 14635254 Review. - Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer?
Flaus A, Owen-Hughes T. Flaus A, et al. Curr Opin Genet Dev. 2004 Apr;14(2):165-73. doi: 10.1016/j.gde.2004.01.007. Curr Opin Genet Dev. 2004. PMID: 15196463 Review.
Cited by
- Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes.
Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Simic R, et al. EMBO J. 2003 Apr 15;22(8):1846-56. doi: 10.1093/emboj/cdg179. EMBO J. 2003. PMID: 12682017 Free PMC article. - The nucleosome remodeling and deacetylase complex in development and disease.
Basta J, Rauchman M. Basta J, et al. Transl Res. 2015 Jan;165(1):36-47. doi: 10.1016/j.trsl.2014.05.003. Epub 2014 May 10. Transl Res. 2015. PMID: 24880148 Free PMC article. Review. - Concerted action of the PHD, chromo and motor domains regulates the human chromatin remodelling ATPase CHD4.
Morra R, Lee BM, Shaw H, Tuma R, Mancini EJ. Morra R, et al. FEBS Lett. 2012 Jul 30;586(16):2513-21. doi: 10.1016/j.febslet.2012.06.017. Epub 2012 Jun 27. FEBS Lett. 2012. PMID: 22749909 Free PMC article. - Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures.
Dürr H, Flaus A, Owen-Hughes T, Hopfner KP. Dürr H, et al. Nucleic Acids Res. 2006;34(15):4160-7. doi: 10.1093/nar/gkl540. Epub 2006 Aug 25. Nucleic Acids Res. 2006. PMID: 16935875 Free PMC article. Review. - Poly(ADP-ribose)-dependent chromatin unfolding facilitates the association of DNA-binding proteins with DNA at sites of damage.
Smith R, Lebeaupin T, Juhász S, Chapuis C, D'Augustin O, Dutertre S, Burkovics P, Biertümpfel C, Timinszky G, Huet S. Smith R, et al. Nucleic Acids Res. 2019 Dec 2;47(21):11250-11267. doi: 10.1093/nar/gkz820. Nucleic Acids Res. 2019. PMID: 31566235 Free PMC article.
References
- Aalfs J.D., Narlikar,G.J. and Kingston,R.E. (2001) Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem., 276, 34270–34278. - PubMed
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. - PubMed
- Akhtar A., Zink,D. and Becker,P.B. (2000) Chromodomains are protein–RNA interaction modules. Nature, 407, 405–409. - PubMed
- Ballestar E., Pile,L.A., Wassarman,D.A., Wolffe,A.P. and Wade,P.A. (2001) A Drosophila MBD family member is a transcriptional corepressor associated with specific genes. Eur. J. Biochem., 268, 5397–5406. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases