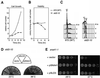SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast - PubMed (original) (raw)
SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast
Reiko Nakajima et al. Mol Biol Cell. 2002 May.
Free PMC article
Abstract
Initiation of DNA replication in eukaryotic cells is regulated through the ordered assembly of replication complexes at origins of replication. Association of Cdc45 with the origins is a crucial step in assembly of the replication machinery, hence can be considered a target for the regulation of origin activation. To examine the process required for SpCdc45 loading, we isolated fission yeast SpSld3, a counterpart of budding yeast Sld3 that interacts with Cdc45. SpSld3 associates with the replication origin during G1-S phases and this association depends on Dbf4-dependent (DDK) kinase activity. In the corresponding period, SpSld3 interacts with minichromosome maintenance (MCM) proteins and then with SpCdc45. A temperature-sensitive sld3-10 mutation suppressed by the multicopy of the sna41+ encoding SpCdc45 impairs loading of SpCdc45 onto chromatin. In addition, this mutation leads to dissociation of preloaded Cdc45 from chromatin in the hydroxyurea-arrested S phase, and DNA replication upon removal of hydroxyurea is retarded. Thus, we conclude that SpSld3 is required for stable association of Cdc45 with chromatin both in initiation and elongation of DNA replication. The DDK-dependent origin association suggests that SpSld3 is involved in temporal regulation of origin firing.
Figures
Figure 1
Structure of S. pombe SpSld3. (A) Schematic representation of SpSld3 is shown. The shaded boxes indicate the coiled-coil regions predicted by COILS program. Asterisks indicate the positions of amino acids changed by the sld3-10,-41, and -52 mutations. (B) Predicted amino acid sequences are aligned with those of S. cerevisiae Sld3, by using the ClustalW method. Identical amino acids are shaded and similar amino acids are hatched by boxshade. Asterisks indicate the positions of amino acids altered by the_sld3-10_, -41, and _-52_mutations as described in the text.
Figure 2
Temperature-sensitive growth of the sld3-10. Exponentially growing wild-type (972) and the sld3-10 cells at 23°C were shifted to 36°C and cells collected at indicated time points were analyzed. The cell number (A) and the cell viability (B) are shown. (C) Flow cytometry profiles are shown. (D) Suppression of sld3-10 by elevated gene dosage of sna41 + is shown. The sld3-10 cells transformed with a vector, pSLD3, or pSNA41 plasmid were streaked out on EMM2 plates and incubated at 23 or 36°C. (E) Suppression of sna41-1 by_sld3_ + is shown. The logarithmically growing sna41-1 cells carrying a vector, pSNA41, or pSLD3 plasmid were spotted after serial dilutions and incubated at 25 or 37°C.
Figure 3
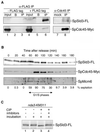
Coimmunoprecipitation of SpCdc45 and SpMcm6 with SpSld3 in G1–S phases. (A) Cells expressing both SpSld3-FLAG5 and SpCdc45-Myc9 (lanes 4–6 and 7 and 8) or only SpCdc45-Myc9 (lanes 1–3) were cultured in the presence of 10 mM HU for 3 h at 30°C to arrest in the early S phase. Whole cell extracts (lanes 1 and 4) were prepared and subjected to immunoprecipitation with anti-FLAG antibody (lanes 3 and 6), mock treatment without antibody (lanes 2 and 5), with normal rabbit serum (lane 7), or anti-SpCdc45 antibody (lane 8). Ten times more immunoprecipitates compared with the input were used for Western blotting with anti-FLAG and anti-Myc antibodies. We did not detect SpSld3-FLAG without immunoprecipitation, probably because the amount was below detection limits. (B) cdc25-22 cells expressing SpSld3-FLAG5 and SpCdc45-Myc9 were arrested for 4 h at 36°C and then released at 25°C. At the indicated time points, extracts were immunoprecipitated with anti-FLAG antibody. SpSld3-FLAG, SpMcm6, and SpCdc45 in the immunoprecipitates were analyzed by immunoblotting. Cell cycle progression was monitored by the percentage of cells containing a septum (septation index), as shown below the panels. In fission yeast, cytokinesis occurs about one-quarter of cell cycle later than nuclear division (Alfa et al., 1993). Thus, the septation index becomes maximum approximately at G1–S transition in wild-type cells, under the conditions we used. (C) SpSld3 was immunoprecipitated using an anti-FLAG antibody from the nda3-KM311 sld3-flag5 cells cultured for 4 h at 20°C (lanes 1–4). Immunoprecipitates were treated with CIP in the presence (lanes 4) or the absence (lanes 3) of phosphatase inhibitors.
Figure 4
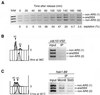
SpSld3 associates with origin DNA in G1–S phases.(A) The cdc25-22 cells expressing SpSld3-FLAG5 and SpCdc45-Myc9 were synchronized, as described for Figure 3B. At every 20 min, cells were fixed with formaldehyde and DNA fragments associated with SpSld3-FLAG were immunoprecipitated with anti-FLAG antibody. The ars2004 and non-ARS regions were PCR amplified from the total cellular DNA (Total) and the immunoprecipitated DNA by using the ars2004 and two non-ARS primer sets and subjected to agarose gel electrophoresis. The percentages of cells containing a septum are shown below the panel. (B) cdc10-V50 sld3-flag5 cells grown at 23°C were incubated for 3 h at 36°C. Cells were subjected to chromatin-immunoprecipitation with anti-FLAG antibody as described above. The flow cytometry profiles at 0 and 3 h are shown (left). (C) hsk1-89 sld3-flag5 cells were shifted to 30°C and incubated for 3 h. Cells were subjected to chromatin-immunoprecipitation with anti-SpMcm6 antibody or anti-FLAG antibody. The flow cytometry profile shows that about half of cells contained unreplicated DNA (left).
Figure 5
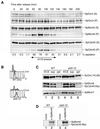
Association of SpCdc45 with chromatin is reduced in sld3-10 mutant. (A) Cell cycle-dependent association of SpCdc45 with chromatin is shown. The_cdc25-22_ cells expressing SpSld3-FLAG5 and SpCdc45-Myc9 were synchronized, as described for Figure 3B. Cells collected at the indicated times were subjected to chromatin fractionation. The supernatant (S) and chromatin-enriched fraction (P) were analyzed by Western blotting with anti-SpMcm6, anti-SpCdc45, and anti-SpOrc4 antibodies. The septation index is shown below the panels. (B) Wild-type and sld3-10 cells expressing SpCdc45-Myc9 and SpOrc1-FLAG5 were grown to log phase at 23°C. The_sld3-10_ cells were then incubated at 36°C for 3 h. The wild-type cells were incubated for 3 h at 36°C in the presence of 12 mM HU to arrest in the early S phase. Results of flow cytometry are shown. (C) Association of SpCdc45 with chromatin in_sld3-10_ was examined. Lysates were subjected to chromatin fractionation. Whole cell extracts (WCE), supernatant (sup), and chromatin-enriched fraction (ppt) were analyzed by immunoblotting with anti-FLAG (top), anti-SpMcm6 and anti-Myc (middle), and anti-Mcm7 (bottom) antibodies. (D) Association of SpMcm6 with SpCdc45 in sld3-10 at restrictive temperature was examined. The wild-type and _sld3-10_cells were incubated at 36°C as in B. Cell lysates were immunoprecipitated with normal rabbit serum (mock IP) or anti-SpMcm6 antibody. Immunoprecipitates were analyzed by immunoblotting with anti-SpMcm6 and anti-Myc antibodies.
Figure 6
SpCdc45 is dissociated from the S-phase–arrested chromatin in sld3-10. (A) Wild-type and_sld3-10_ cells expressing SpOrc1-FLAG5 and SpCdc45-Myc9 were cultured for 4 h in the presence of 10 mM HU at the permissive temperature (23°C) and then incubated at 36°C for 1 h. Cells were washed and resuspended into the complete medium without HU. (A) Scheme of the experiment is shown. (B) DNA contents of cells after release from HU arrest were analyzed by flow cytometry. The cell numbers (C) and the viability obtained by colony-forming units/cell number (D) are shown. (E) Chromatin fraction was prepared from the HU-arrested cells before and after the incubation for 1 h at 36°C. Whole cell extracts (W), supernatant (S), and chromatin-enriched fraction (P) were analyzed by immunoblotting with anti-FLAG for SpOrc1-FL (top), anti-SpMcm6 (middle), and anti-Myc antibodies for SpCdc45-Myc (bottom). (F) Images in E were quantitated using the LAS1000 plus image analyzer (Fuji Film, Tokyo, Japan) and ratios of SpMcm6/SpOrc1 and SpCdc45/SpOrc1 are presented.
Similar articles
- Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae.
Kamimura Y, Tak YS, Sugino A, Araki H. Kamimura Y, et al. EMBO J. 2001 Apr 17;20(8):2097-107. doi: 10.1093/emboj/20.8.2097. EMBO J. 2001. PMID: 11296242 Free PMC article. - Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins.
Yabuuchi H, Yamada Y, Uchida T, Sunathvanichkul T, Nakagawa T, Masukata H. Yabuuchi H, et al. EMBO J. 2006 Oct 4;25(19):4663-74. doi: 10.1038/sj.emboj.7601347. Epub 2006 Sep 21. EMBO J. 2006. PMID: 16990792 Free PMC article. - Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing.
Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Tanaka S, et al. Curr Biol. 2011 Dec 20;21(24):2055-63. doi: 10.1016/j.cub.2011.11.038. Epub 2011 Dec 8. Curr Biol. 2011. PMID: 22169533 - Origins and complexes: the initiation of DNA replication.
Bryant JA, Moore K, Aves SJ. Bryant JA, et al. J Exp Bot. 2001 Feb;52(355):193-202. J Exp Bot. 2001. PMID: 11283163 Review. - Cdc6 and DNA replication: limited to humble origins.
Heichman KA. Heichman KA. Bioessays. 1996 Nov;18(11):859-62. doi: 10.1002/bies.950181103. Bioessays. 1996. PMID: 8939063 Review.
Cited by
- Evolutionary conservation of the CDK targets in eukaryotic DNA replication initiation.
Zegerman P. Zegerman P. Chromosoma. 2015 Sep;124(3):309-21. doi: 10.1007/s00412-014-0500-y. Epub 2015 Jan 11. Chromosoma. 2015. PMID: 25575982 Review. - GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication.
Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V. Balestrini A, et al. Nat Cell Biol. 2010 May;12(5):484-91. doi: 10.1038/ncb2050. Epub 2010 Apr 11. Nat Cell Biol. 2010. PMID: 20383140 Free PMC article. - Activation of the DNA damage checkpoint in mutants defective in DNA replication initiation.
Yin L, Locovei AM, D'Urso G. Yin L, et al. Mol Biol Cell. 2008 Oct;19(10):4374-82. doi: 10.1091/mbc.e08-01-0020. Epub 2008 Jul 30. Mol Biol Cell. 2008. PMID: 18667534 Free PMC article. - Schizosaccharomyces pombe replication protein Cdc45/Sna41 requires Hsk1/Cdc7 and Rad4/Cut5 for chromatin binding.
Dolan WP, Sherman DA, Forsburg SL. Dolan WP, et al. Chromosoma. 2004 Sep;113(3):145-56. doi: 10.1007/s00412-004-0302-8. Epub 2004 Aug 3. Chromosoma. 2004. PMID: 15338237 - Eukaryotic MCM proteins: beyond replication initiation.
Forsburg SL. Forsburg SL. Microbiol Mol Biol Rev. 2004 Mar;68(1):109-31. doi: 10.1128/MMBR.68.1.109-131.2004. Microbiol Mol Biol Rev. 2004. PMID: 15007098 Free PMC article. Review.
References
- Alfa C, Fantes P, Hyams J, Mcleod M, Warbrick E. Experiments with Fission Yeast, A Laboratory Course Manual. Cold Spring Harbor, NY.: Cold Spring Harbor Laboratory Press; 1993.
- Arata Y, Fujita M, Ohtani K, Kijima S, Kato JY. Cdk2-dependent and -independent pathways in E2F-mediated S phase induction. J Biol Chem. 2000;275:6337–6345. - PubMed
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases